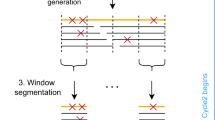
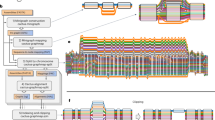
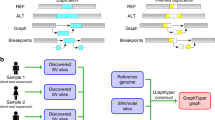
Abstract
Reference genomes guide our interpretation of DNA sequence data. However, conventional linear references represent only one version of each locus, ignoring variation in the population. Poor representation of an individual′s genome sequence impacts read mapping and introduces bias. Variation graphs are bidirected DNA sequence graphs that compactly represent genetic variation across a population, including large-scale structural variation such as inversions and duplications1. Previous graph genome software implementations2,3,4 have been limited by scalability or topological constraints. Here we present vg, a toolkit of computational methods for creating, manipulating, and using these structures as references at the scale of the human genome. vg provides an efficient approach to mapping reads onto arbitrary variation graphs using generalized compressed suffix arrays5, with improved accuracy over alignment to a linear reference, and effectively removing reference bias. These capabilities make using variation graphs as references for DNA sequencing practical at a gigabase scale, or at the topological complexity of de novo assemblies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Paten, B., Novak, A.M., Eizenga, J.M. & Garrison, E. Genome graphs and the evolution of genome inference. Genome Res. 27, 665–676 (2017).
Dilthey, A., Cox, C., Iqbal, Z., Nelson, M.R. & McVean, G. Improved genome inference in the MHC using a population reference graph. Nat. Genet. 47, 682–688 (2015).
Eggertsson, H.P. et al. Graphtyper enables population-scale genotyping using pangenome graphs. Nat. Genet. 49, 1654–1660 (2017).
Rakocevic, G. et al. Fast and accurate genomic analyses using genome graphs. Preprint @bioRxiv https://doi.org/10.1101/194530 (2017).
Siren, J. Indexing variation graphs. Proc. 19th Workshop on Algorithm Engineering and Experiments (ALENEX) (Society for Industrial and Applied Mathematics, 2017).
Delcher, A.L. et al. Alignment of whole genomes. Nucleic Acids Res. 27, 2369–2376 (1999).
Paten, B. et al. Cactus: Algorithms for genome multiple sequence alignment. Genome Res. 21, 1512–1528 (2011).
Li, R. et al. Building the sequence map of the human pan-genome. Nat. Biotechnol. 28, 57–63 (2010).
Yuan, S. & Qin, Z. Read-mapping using personalized diploid reference genome for RNA sequencing data reduced bias for detecting allele specific expression. IEEE International Conference on Bioinformatics and Biomedicine Workshops (IEEE, 2012).
Lee, C., Grasso, C. & Sharlow, M.F. Multiple sequence alignment using partial order graphs. Bioinformatics 18, 452–464 (2002).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Pevzner, P.A., Tang, H. & Waterman, M.S. An Eulerian path approach to DNA fragment assembly. Proc. Natl. Acad. Sci. USA 98, 9748–9753 (2001).
Myers, E.W. The fragment assembly string graph. Bioinformatics 21 (Suppl. 2), ii79–ii85 (2005).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint @ https://doi.org/arxiv.org/abs/1207.3907 (2012).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Novak, A.M. et al. Genome graphs. Preprint @ bioRxiv https://doi.org/10.1101/101378 (2017).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with bwa-mem. Preprint @ https://doi.org/arxiv.org/abs/1303.3997 (2013).
Zook, J.M. et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data 3, 160025 (2016).
McDaniell, R. et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328, 235–239 (2010).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Yue, J.-X. et al. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat. Genet. 49, 913–924 (2017).
Aguirre de Cárcer, D., López-Bueno, A., Pearce, D.A. & Alcamí, A. Biodiversity and distribution of polar freshwater DNA viruses. Sci. Adv. 1, e1400127 (2015).
Chikhi, R. & Rizk, G. Space-efficient and exact de Bruijn graph representation based on a Bloom filter. Algorithms Mol. Biol. 8, 22 (2013).
Durbin, R. Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT). Bioinformatics 30, 1266–1272 (2014).
Novak, A.M., Garrison, E. & Paten, B. in Algorithms in Bioinformatics (eds. Firth, M. & Pedersen, C.N.) 246–256 (Springer, Heidelberg, 2016).
Ge, B. et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 41, 1216–1222 (2009).
Beretta, S. et al. in Algorithms for Computational Biology (AlCoB) 2017, (eds. Figueiredo, D., Martn-Vide, C., Pratas, D. & Vega-Rodrguez, M.) 49–61 Lecture Notes in Computer Science 10252 (Springer, Champaign-Urbana, 2017).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Gog, S., Beller, T., Moat, A. & Petri, M. in International Symposium on Experimental Algorithms 326–337 (Springer, 2014).
Myers, E.W. & Miller, W. Approximate matching of regular expressions. Bull. Math. Biol. 51, 5–37 (1989).
Farrar, M. Striped Smith-Waterman speeds database searches six times over other SIMD implementations. Bioinformatics 23, 156–161 (2007).
Durbin, R., Eddy, S.R., Krogh, A. & Mitchison, G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids (Cambridge University Press, 1998).
Hamada, M., Wijaya, E., Frith, M.C. & Asai, K. Probabilistic alignments with quality scores: an application to short-read mapping toward accurate SNP/indel detection. Bioinformatics 27, 3085–3092 (2011).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011).
Acknowledgements
E.G., J.S., and R.D. were funded by the Wellcome Trust (grants 206194 and 207492). E.T.D. was funded by an NIH Cambridge Trust studentship, and W.J. by a Wellcome Trust MGM studentship (109083/Z/15/Z). A.M.N., G.H., J.M.E., and B.P. were supported by the National Institutes of Health (5U41HG007234), the W.M. Keck Foundation (DT06172015) and the Simons Foundation (SFLIFE# 35190). We thank members of the GA4GH Reference Variation Working Group for support, ideas, and comments, and Hannes Eggertsson for assistance in the integration with GraphTyper.
Author information
Authors and Affiliations
Contributions
E.G. conceived and led the development of vg, J.S. developed the GCSA2 index, A.M.N., G.H., J.M.E., and E.T.D. contributed to the software, E.G., W.J., S.G., C.M., M.F.L., and R.D. contributed results and data analysis, R.D. and B.P. oversaw the project, and all contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.L. is an employee of, and E.G. consults for, DNAnexus Inc. R.D. holds shares in and consults for Congenica Ltd. and Dovetail Inc.
Integrated supplementary information
Supplementary Figure 1 ROC curves as in main figure 2a for vg graphs with different allele frequency inclusion thresholds.
ROC curves parameterised by mapping quality for 10M read pairs simulated from NA24385 as mapped by bwa mem, vg to various 1000GP pangenome references, and vg with a linear reference, using single end (se) or paired end (pe) mapping. Allele frequency thresholds for the various rows are from top to bottom 0 (all variants), 0.001, 0.01, 0.1. Within each row, the left plot is based on all reads, middle on reads simulated from segments with no genetic variants from the linear reference, right on reads simulated from segments containing variants. All reads may contain simulated sequencing errors.
Supplementary Figure 2 Relative performance of vg and bwa mem when mapping to a viral metagenome assembly graph.
Left: part of the assembly graph for an arctic viral metagenome23 assembled by minia and visualized by Bandage after complexity reduction in vg. Right: a scatterplot showing the score obtained when aligning each of 100,000 reads held out from the metagenomic assembly with bwa mem to the contigs of the assembly (y-axis) versus vg to the assembly graph (x-axis).
Supplementary Figure 3 Illustration of the unfolding process.
The starting graph (a) has an inverting edge leading from the forward to reverse strand of node 2. In (b) we unfold the graph and unroll with k greater than the length of the graph, which materializes the implied reverse strand as sequence on the forward strand of new nodes.
Supplementary Figure 4 Illustration of the unrolling process.
The starting graph (a) and a representation without sequences or sides to clarify the underlying structure (b). In (c) we have unrolled one step (k = 2). In (d), k = 4, (e) k = 10, and (f) k = 25.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 842 kb)
Supplementary Information
Supplementary Table 1, Supplementary Note, and Supplementary References (PDF 382 kb)
Rights and permissions
About this article
Cite this article
Garrison, E., Sirén, J., Novak, A. et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat Biotechnol 36, 875–879 (2018). https://doi.org/10.1038/nbt.4227
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4227
This article is cited by
-
Amplidiff: an optimized amplicon sequencing approach to estimating lineage abundances in viral metagenomes
BMC Bioinformatics (2024)
-
Introgressions lead to reference bias in wheat RNA-seq analysis
BMC Biology (2024)
-
Co-linear chaining on pangenome graphs
Algorithms for Molecular Biology (2024)
-
A comprehensive benchmark of graph-based genetic variant genotyping algorithms on plant genomes for creating an accurate ensemble pipeline
Genome Biology (2024)
-
Suffix sorting via matching statistics
Algorithms for Molecular Biology (2024)