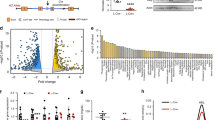
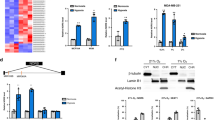
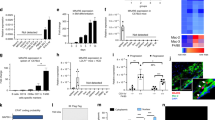
Abstract
Nuclear receptors regulate gene expression in response to environmental cues, but the molecular events governing the cell type specificity of nuclear receptors remain poorly understood. Here we outline a role for a long noncoding RNA (lncRNA) in modulating the cell type–specific actions of liver X receptors (LXRs), sterol-activated nuclear receptors that regulate the expression of genes involved in cholesterol homeostasis and that have been causally linked to the pathogenesis of atherosclerosis. We identify the lncRNA MeXis as an amplifier of LXR-dependent transcription of the gene Abca1, which is critical for regulation of cholesterol efflux. Mice lacking the MeXis gene show reduced Abca1 expression in a tissue-selective manner. Furthermore, loss of MeXis in mouse bone marrow cells alters chromosome architecture at the Abca1 locus, impairs cellular responses to cholesterol overload, and accelerates the development of atherosclerosis. Mechanistic studies reveal that MeXis interacts with and guides promoter binding of the transcriptional coactivator DDX17. The identification of MeXis as a lncRNA modulator of LXR-dependent gene expression expands understanding of the mechanisms underlying cell type–selective actions of nuclear receptors in physiology and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rohatgi, A. et al. HDL cholesterol efflux capacity and incident cardiovascular events. N. Engl. J. Med. 371, 2383–2393 (2014).
de la Llera-Moya, M. et al. The ability to promote efflux via ABCA1 determines the capacity of serum specimens with similar high-density lipoprotein cholesterol to remove cholesterol from macrophages. Arterioscler. Thromb. Vasc. Biol. 30, 796–801 (2010).
Libby, P. et al. Inflammation in atherosclerosis: from pathophysiology to practice. J. Am. Coll. Cardiol. 54, 2129–2138 (2009).
Cao, Q. et al. The central role of EED in the orchestration of polycomb group complexes. Nat. Commun. 5, 3127 (2014).
Zelcer, N. & Tontonoz, P. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Invest. 116, 607–614 (2006).
Bodzioch, M. et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 22, 347–351 (1999).
Rust, S. et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 22, 352–355 (1999).
Rinn, J.L. & Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Wang, K.C. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011).
Kotzin, J.J. et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537, 239–243 (2016).
Engreitz, J.M. et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016).
Zhang, L. et al. Inhibition of cholesterol biosynthesis through RNF145-dependent ubiquitination of SCAP. eLife 6, e28766 (2017).
Sallam, T. et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128 (2016).
Creyghton, M.P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936 (2010).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Quinn, J.J. & Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 (2016).
Huarte, M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010).
Cheng, J.H., Pan, D.Z., Tsai, Z.T. & Tsai, H.K. Genome-wide analysis of enhancer RNA in gene regulation across 12 mouse tissues. Sci. Rep. 5, 12648 (2015).
Daniel, B. et al. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev. 28, 1562–1577 (2014).
Chu, C. et al. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015).
Auboeuf, D., Hönig, A., Berget, S.M. & O'Malley, B.W. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298, 416–419 (2002).
Wortham, N.C. et al. The DEAD-box protein p72 regulates ERα-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERα-positive breast cancer. Oncogene 28, 4053–4064 (2009).
Nikpay, M. et al. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013).
Wang, K.C. & Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011).
Khalil, A.M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 106, 11667–11672 (2009).
Sallam, T. et al. The macrophage LBP gene is an LXR target that promotes macrophage survival and atherosclerosis. J. Lipid Res. 55, 1120–1130 (2014).
Rong, X. et al. LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 18, 685–697 (2013).
Jakobsson, T. et al. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Mol. Cell 34, 510–518 (2009).
Wongpalee, S.P. et al. Large-scale remodeling of a repressed exon ribonucleoprotein to an exon definition complex active for splicing. eLife 5, e19743 (2016).
Shin, K.J. et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. Sci. USA 103, 13759–13764 (2006).
Boyden, E.S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005).
Peet, D.J. et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell 93, 693–704 (1998).
Ito, A. et al. LXRs link metabolism to inflammation through Abca1-dependent regulation of membrane composition and TLR signaling. eLife 4, e08009 (2015).
Bhatt, D.M. et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150, 279–290 (2012).
Bradley, M.N. et al. Ligand activation of LXRβ reverses atherosclerosis and cellular cholesterol overload in mice lacking LXRα and apoE. J. Clin. Invest. 117, 2337–2346 (2007).
Tangirala, R.K., Rubin, E.M. & Palinski, W. Quantitation of atherosclerosis in murine models: correlation between lesions in the aortic origin and in the entire aorta, and differences in the extent of lesions between sexes in LDL receptor–deficient and apolipoprotein E–deficient mice. J. Lipid Res. 36, 2320–2328 (1995).
Feig, J.E. & Fisher, E.A. Laser capture microdissection for analysis of macrophage gene expression from atherosclerotic lesions. Methods Mol. Biol. 1027, 123–135 (2013).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Zhao, Y. et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 44 (D1), D203–D208 (2016).
Koeth, R.A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Sun, L. et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41, e166 (2013).
McLean, C.Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Tsai, M.C. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Buenrostro, J.D., Giresi, P.G., Zaba, L.C., Chang, H.Y. & Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Soneson, C., Love, M.I. & Robinson, M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Acknowledgements
We thank members of the Tontonoz, Smale, Black, and Nagy laboratories and the University of California Los Angeles (UCLA) Atherosclerosis Research Unit for technical assistance and useful discussions. This work was supported by the National Institutes of Health grants HL030568, HL066088, HL128822, the Burroughs Wellcome Fund Career Award for Medical Scientists, and the UCLA Cardiovascular Discovery Fund (Lauren B. Leichtman and Arthur E. Levine Investigator Award).
Author information
Authors and Affiliations
Contributions
T.S. and P.T. conceived and designed the study and guided the interpretation of the results and the preparation of the manuscript. P.T. supervised the study, and T.S. managed the daily experiments. X.W. performed most mouse experiments and data analysis, including for the atherosclerosis study. T.S., M.J., T.G., K.Q., Z.Z., J.S., D.S., P.R., C.H., A.I., and X.L. performed various in vivo and in vitro macrophage experiments and data analysis. Bioinformatic data analysis was performed by D.C. and A.E. B.J.T., X.L., and S.S. assisted with ChIP and ATAC-seq experiments, including data analysis. J.W. performed the mass spectrometry analysis. B.D. and L.N. assisted with ChIP and performed experiments defining enhancer landscape at Abca1. A.C. performed ChIP–seq for LXR. M.C. and A.J.L. provided GWAS data and technical guidance for atherosclerosis analysis. T.S. and P.T. edited the manuscript with input from all authors. All authors discussed the results and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures & Tables
Supplementary Figures 1–14 & Supplementary Tables 1,3–4 (PDF 6140 kb)
Supplementary Table 2
Analysis of lncRNA-adjacent protein-coding genes (XLSX 117 kb)
Rights and permissions
About this article
Cite this article
Sallam, T., Jones, M., Thomas, B. et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med 24, 304–312 (2018). https://doi.org/10.1038/nm.4479
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4479
This article is cited by
-
The roles and molecular mechanisms of non-coding RNA in cancer metabolic reprogramming
Cancer Cell International (2024)
-
Elder (Sambucus nigra), identified by high-content screening, counteracts foam cell formation without promoting hepatic lipogenesis
Scientific Reports (2024)
-
Macrophage-based therapeutic approaches for cardiovascular diseases
Basic Research in Cardiology (2024)
-
Integrative regulation of hLMR1 by dietary and genetic factors in nonalcoholic fatty liver disease and hyperlipidemia
Human Genetics (2024)
-
SNHG17 alters anaerobic glycolysis by resetting phosphorylation modification of PGK1 to foster pro-tumor macrophage formation in pancreatic ductal adenocarcinoma
Journal of Experimental & Clinical Cancer Research (2023)