Abstract
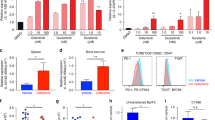
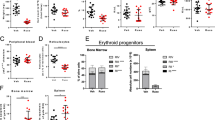
Individuals with acute myeloid leukemia (AML) harboring an internal tandem duplication (ITD) in the gene encoding Fms-related tyrosine kinase 3 (FLT3) who relapse after allogeneic hematopoietic cell transplantation (allo-HCT) have a 1-year survival rate below 20%. We observed that sorafenib, a multitargeted tyrosine kinase inhibitor, increased IL-15 production by FLT3-ITD+ leukemia cells. This synergized with the allogeneic CD8+ T cell response, leading to long-term survival in six mouse models of FLT3-ITD+ AML. Sorafenib-related IL-15 production caused an increase in CD8+CD107a+IFN-γ+ T cells with features of longevity (high levels of Bcl-2 and reduced PD-1 levels), which eradicated leukemia in secondary recipients. Mechanistically, sorafenib reduced expression of the transcription factor ATF4, thereby blocking negative regulation of interferon regulatory factor 7 (IRF7) activation, which enhanced IL-15 transcription. Both IRF7 knockdown and ATF4 overexpression in leukemia cells antagonized sorafenib-induced IL-15 production in vitro. Human FLT3-ITD+ AML cells obtained from sorafenib responders following sorafenib therapy showed increased levels of IL-15, phosphorylated IRF7, and a transcriptionally active IRF7 chromatin state. The mitochondrial spare respiratory capacity and glycolytic capacity of CD8+ T cells increased upon sorafenib treatment in sorafenib responders but not in nonresponders. Our findings indicate that the synergism of T cells and sorafenib is mediated via reduced ATF4 expression, causing activation of the IRF7–IL-15 axis in leukemia cells and thereby leading to metabolic reprogramming of leukemia-reactive T cells in humans. Therefore, sorafenib treatment has the potential to contribute to an immune-mediated cure of FLT3-ITD-mutant AML relapse, an otherwise fatal complication after allo-HCT.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
06 March 2018
In the version of this article initially published, Omid Shah’s name was misspelled as Omid Sha. The error has been corrected in the PDF and HTML versions of this article.
References
Pfirrmann, M. et al. Prediction of post-remission survival in acute myeloid leukaemia: a post-hoc analysis of the AML96 trial. Lancet Oncol. 13, 207–214 (2012).
Sengsayadeth, S.M. et al. Allo-SCT for high-risk AML-CR1 in the molecular era: impact of FLT3/ITD outweighs the conventional markers. Bone Marrow Transplant. 47, 1535–1537 (2012).
Serve, H. et al. Sorafenib in combination with intensive chemotherapy in elderly patients with acute myeloid leukemia: results from a randomized, placebo-controlled trial. J. Clin. Oncol. 31, 3110–3118 (2013).
Röllig, C. et al. Addition of sorafenib versus placebo to standard therapy in patients aged 60 years or younger with newly diagnosed acute myeloid leukaemia (SORAML): a multicentre, phase 2, randomised controlled trial. Lancet Oncol. 16, 1691–1699 (2015).
Metzelder, S.K. et al. High activity of sorafenib in FLT3-ITD-positive acute myeloid leukemia synergizes with allo-immune effects to induce sustained responses. Leukemia 26, 2353–2359 (2012).
Tschan-Plessl, A. et al. Synergistic effect of sorafenib and cGvHD in patients with high-risk FLT3-ITD+ AML allows long-term disease control after allogeneic transplantation. Ann. Hematol. 94, 1899–1905 (2015).
Krüger, W.H. et al. Molecular remission of FLT3-ITD+ AML relapse after allo-SCT by acute GVHD in addition to sorafenib. Bone Marrow Transplant. 47, 137–138 (2012).
Chen, Y.B. et al. Phase I trial of maintenance sorafenib after allogeneic hematopoietic stem cell transplantation for Fms-like tyrosine kinase 3 internal tandem duplication acute myeloid leukemia. Biol. Blood Marrow Transplant. 20, 2042–2048 (2014).
Antar, A., Kharfan-Dabaja, M.A., Mahfouz, R. & Bazarbachi, A. Sorafenib maintenance appears safe and improves clinical outcomes in FLT3-ITD acute myeloid leukemia after allogeneic hematopoietic cell transplantation. Clin. Lymphoma Myeloma Leuk. 15, 298–302 (2015).
Tarlock, K. et al. Sorafenib treatment following hematopoietic stem cell transplant in pediatric FLT3/ITD acute myeloid leukemia. Pediatr. Blood Cancer 62, 1048–1054 (2015).
Brunner, A.M. et al. Haematopoietic cell transplantation with and without sorafenib maintenance for patients with FLT3-ITD acute myeloid leukaemia in first complete remission. Br. J. Haematol. 175, 496–504 (2016).
Zorko, N.A. et al. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood 120, 1130–1136 (2012).
Bernot, K.M. et al. Eradicating acute myeloid leukemia in a MllPTD/wt:Flt3ITD/wt murine model: a path to novel therapeutic approaches for human disease. Blood 122, 3778–3783 (2013).
Lucas, M., Schachterle, W., Oberle, K., Aichele, P. & Diefenbach, A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity 26, 503–517 (2007).
Rubio, V. et al. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat. Med. 9, 1377–1382 (2003).
Pandiyan, P. et al. The role of IL-15 in activating STAT5 and fine-tuning IL-17A production in CD4 T lymphocytes. J. Immunol. 189, 4237–4246 (2012).
Patsoukis, N. et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692–6697 (2015).
van Bockel, D.J. et al. Persistent survival of prevalent clonotypes within an immunodominant HIV gag-specific CD8+ T cell response. J. Immunol. 186, 359–371 (2011).
Azimi, N., Shiramizu, K.M., Tagaya, Y., Mariner, J. & Waldmann, T.A. Viral activation of interleukin-15 (IL-15): characterization of a virus-inducible element in the IL-15 promoter region. J. Virol. 74, 7338–7348 (2000).
Romieu-Mourez, R. et al. Distinct roles for IFN regulatory factor (IRF)-3 and IRF-7 in the activation of antitumor properties of human macrophages. Cancer Res. 66, 10576–10585 (2006).
Liang, Q., Deng, H., Sun, C.W., Townes, T.M. & Zhu, F. Negative regulation of IRF7 activation by activating transcription factor 4 suggests a cross-regulation between the IFN responses and the cellular integrated stress responses. J. Immunol. 186, 1001–1010 (2011).
Smith, C.C., Lin, K., Stecula, A., Sali, A. & Shah, N.P. FLT3 D835 mutations confer differential resistance to type II FLT3 inhibitors. Leukemia 29, 2390–2392 (2015).
Buck, M.D., O'Sullivan, D. & Pearce, E.L. T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015).
van der Windt, G.J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
van der Windt, G.J. et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl. Acad. Sci. USA 110, 14336–14341 (2013).
Schultze-Florey, C. et al. TCR diversity is a predictive marker for donor lymphocyte infusion response. Blood 128, 4605 (2016).
van Bergen, C.A. et al. Selective graft-versus-leukemia depends on magnitude and diversity of the alloreactive T cell response. J. Clin. Invest. 127, 517–529 (2017).
Thiant, S. et al. Plasma levels of IL-7 and IL-15 in the first month after myeloablative BMT are predictive biomarkers of both acute GVHD and relapse. Bone Marrow Transplant. 45, 1546–1552 (2010).
Sauter, C.T. et al. Interleukin-15 administration increases graft-versus-tumor activity in recipients of haploidentical hematopoietic SCT. Bone Marrow Transplant. 48, 1237–1242 (2013).
Chen, G. et al. Expanded donor natural killer cell and IL-2, IL-15 treatment efficacy in allogeneic hematopoietic stem cell transplantation. Eur. J. Haematol. 81, 226–235 (2008).
Blaser, B.W. et al. Donor-derived IL-15 is critical for acute allogeneic graft-versus-host disease. Blood 105, 894–901 (2005).
Blaser, B.W. et al. Trans-presentation of donor-derived interleukin 15 is necessary for the rapid onset of acute graft-versus-host disease but not for graft-versus-tumor activity. Blood 108, 2463–2469 (2006).
Roychowdhury, S. et al. IL-15 but not IL-2 rapidly induces lethal xenogeneic graft-versus-host disease. Blood 106, 2433–2435 (2005).
Cario, G. et al. High interleukin-15 expression characterizes childhood acute lymphoblastic leukemia with involvement of the CNS. J. Clin. Oncol. 25, 4813–4820 (2007).
Wu, S. et al. Expression of interleukin 15 in primary adult acute lymphoblastic leukemia. Cancer 116, 387–392 (2010).
Nasilowska-Adamska, B. et al. Mild chronic graft-versus-host disease may alleviate poor prognosis associated with FLT3 internal tandem duplication for adult acute myeloid leukemia following allogeneic stem cell transplantation with myeloablative conditioning in first complete remission: a retrospective study. Eur. J. Haematol. 96, 236–244 (2016).
Barnes, B.J., Moore, P.A. & Pitha, P.M. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon α genes. J. Biol. Chem. 276, 23382–23390 (2001).
Steinmann, J. et al. 5-Azacytidine and DLI can induce long-term remissions in AML patients relapsed after allograft. Bone Marrow Transplant. 50, 690–695 (2015).
Bejanyan, N. et al. Survival of patients with acute myeloid leukemia relapsing after allogeneic hematopoietic cell transplantation: a center for international blood and marrow transplant research study. Biol. Blood Marrow Transplant. 21, 454–459 (2015).
Takami, A. et al. Donor lymphocyte infusion for the treatment of relapsed acute myeloid leukemia after allogeneic hematopoietic stem cell transplantation: a retrospective analysis by the Adult Acute Myeloid Leukemia Working Group of the Japan Society for Hematopoietic Cell Transplantation. Biol. Blood Marrow Transplant. 20, 1785–1790 (2014).
Buck, M.D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Bolger, A.M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Boeva, V. et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 (2012).
Carvalho, B.S. & Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010).
Luo, W., Friedman, M.S., Shedden, K., Hankenson, K.D. & Woolf, P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161 (2009).
Kamburov, A., Stelzl, U., Lehrach, H. & Herwig, R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 41, D793–D800 (2013).
Brummer, T. et al. Phosphorylation-dependent binding of 14-3-3 terminates signalling by the Gab2 docking protein. EMBO J. 27, 2305–2316 (2008).
Wilhelm, K. et al. Graft-versus-host disease enhanced by extracellular adenosine triphosphate activating P2X7R. Nat. Med. 16, 1434–1438 (2010).
Schwab, L. et al. Neutrophil granulocytes recruited upon translocation of intestinal bacteria enhance GvHD via tissue damage. Nat. Med. 20, 648–654 (2014).
Kaplan, D.H. et al. Target antigens determine graft-versus-host disease phenotype. J. Immunol. 173, 5467–5475 (2004).
Acknowledgements
We thank G. Prinz and H. Dierbach for their help with mouse experiments, K. Geiger and D. Herchenbach for cell sorting, and S. Decker (University of Freiburg) for providing NSG mice. We thank M.E.D. Flowers (University of Washington) for help with patient data. We thank D. Cittaro for the help with bioinformatic analysis. Il15−/− mice were provided by Y. Tanriver (University of Freiburg). Il15−/− mice were provided by B. Becher (University of Zurich).
This study was supported by the German Research Foundation (DFG) Heisenberg Professorship ZE 872/3-1 (R.Z.), DFG Sonderforschungsbereiche 1074 (SFB1074; F.K.), SFB1160 (R.Z.), SFB850 (T.B.), and TRR167 (R.Z.); European Research Council (ERC) Consolidator Grant no. 681012 GvHDCure (R.Z.); Deutsche Krebshilfe no. 111639 (G.H., R.Z.); Deutsche Jose Carreras Leukämie-Stiftung (DJCLS; G.H., R.Z.); Else Kröner-Fresenius Foundation (EKF) Stiftung no. 2015_A147 (P.A.), INTERREG V Rhin Supérieur (P.A., R.Z.); LOEWE–Gentherapie Frankfurt (CGT), Hessian Ministry of Higher Education, Research and the Arts, Germany no. III L 4- 518/17.004 (E.U.); Max-Eder-Nachwuchsgruppenprogramm, Deutsche Krebshilfe no. 109420 (F.K.); European Hematology Association fellowship 2010/04 (F.K.); and National Institutes of Health (NIH) grant no. R01 CA-72669 (B.R.B.). E.R. was supported by a fellowship from Associazione Italiana per la Ricerca sul Cancro (AIRC) that was cofunded by the European Union.
Author information
Authors and Affiliations
Contributions
N.R.M. performed the majority of the experiments, helped develop the overall concept behind the study, and helped write the manuscript. F.B. helped with the experiments and with development of the overall concept behind the study. L.B. performed ATF4 overexpression experiments. D.O'S. helped with Seahorse analysis. S. Thomas and S. Tugues helped with mouse experiments. M.W., T.A.M., K.H., P.A., A.L.I., G.I., K.S., W.M., S. Duqusne and A.W. helped with experiments and data interpretation. A.S.-G. performed immunohistological analysis. L.O. and K.-L.Y. helped with experiments. D.P., M.F., R. Claus, M. Lübbert, C.R., H. Bertz, R.W., J.H., A. Schmidts, M.S., D. Bettinger, R.T., E.U., Y.T., G.L.V., R.A., P.H., D. Wolf, M.D., C.J., K.W., C. Leiber, S. Gerull, J.H., C. Lengerke, T.P., T.S., G.K., W.R., S. Doostkam, S.M., and S.K.M. provided patient data. S. Taromi, S.S., and B.B. helped with mouse experiments. S.H. and T.B. helped with western blot and knockdown experiments. Z.H. and J. Dengjel performed mass spectrometry and data analysis. S.K. and B.K. performed mass spectrometry of sorafenib binding partners and kinome analysis. B.H., C. Schmid, U.H., C. Scheid, A. Spyridonidis, F.S., R.O., L.P.M., F.S.-d.-F., and J.K. provided patient data and helped with the analysis. M.P. performed analysis of the biopsy specimen. A.B., A. Nagler, D. Bunjes, A.M., W.H., and G.S. provided patient data and helped develop the overall concept behind the study. J.E.E. and D.F. analyzed the level of FLT3 inhibition upon sorafenib exposure. E.-M.W., J.-Y.C., F.K., D. Beelen, R. Chakraverty, S.R., S. Gill, N.K., F.A., L.V., J.S., and F.C. provided and analyzed patient data. E.R. and C.B. performed TRC sequencing and analyzed related data. A.M.M., T.K., T.T., B.K., D.K., D. Weisdorf, W.v.d.V., D.D., W.B., I.H., A.H., G.A., M. Börries, H. Busch, J.M., P.R., M. Labopin, J.H.A., A.S.H., G.R.H., G.A.K., M. Bar, A. Sarma, D.M., G.M., B.O., K.R., O.S., R.S.N., and A. Neubauer provided and analyzed patient data. E.U. and M.A.C. provided reagents and contributed to the development of the concept behind the study and the manuscript. B.R.B., N.v.B., and G.H. provided reagents, helped with the experiments, and analyzed data. E.P. helped to plan and analyze the T cell metabolism experiments. J. Duyster and J.F. helped develop the concept behind the study, analyze the data, and write the manuscript. R.Z. developed the overall concept behind the study, supervised the experiments, analyzed the data, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures & Tables
Supplementary Figures 1–21 & Supplementary Tables 2–12 (PDF 8654 kb)
Supplementary Table 1
Patients raw data (XLSX 69 kb)
Rights and permissions
About this article
Cite this article
Mathew, N., Baumgartner, F., Braun, L. et al. Sorafenib promotes graft-versus-leukemia activity in mice and humans through IL-15 production in FLT3-ITD-mutant leukemia cells. Nat Med 24, 282–291 (2018). https://doi.org/10.1038/nm.4484
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4484
This article is cited by
-
Pretransplant hepatomegaly is linked to relapse in patients with leukemia and myelodysplastic syndrome not in remission
International Journal of Hematology (2024)
-
ROCK1/2 signaling contributes to corticosteroid-refractory acute graft-versus-host disease
Nature Communications (2024)
-
Relapse of acute myeloid leukemia after allogeneic stem cell transplantation: immune escape mechanisms and current implications for therapy
Molecular Cancer (2023)
-
Hypomethylating agents plus modified priming regimens compared with venetoclax-based regimens based on molecular characteristics for newly diagnosed patients with acute myeloid leukemia: a multi-center cohort study
Annals of Hematology (2023)
-
Post-transplant maintenance therapy in acute myeloid leukemia after allogeneic hematopoietic stem cell transplantation harmonizing multiple therapeutic modalities including targeted therapy, immunotherapy and cellular therapy
International Journal of Hematology (2023)