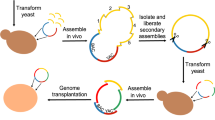
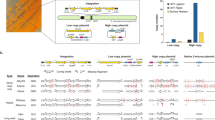
Abstract
Recent exponential advances in genome sequencing and engineering technologies have enabled an unprecedented level of interrogation into the impact of DNA variation (genotype) on cellular function (phenotype). Furthermore, these advances have also prompted realistic discussion of writing and radically re-writing complex genomes. In this Perspective, we detail the motivation for large-scale engineering, discuss the progress made from such projects in bacteria and yeast and describe how various genome-engineering technologies will contribute to this effort. Finally, we describe the features of an ideal platform and provide a roadmap to facilitate the efficient writing of large genomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kosuri, S. & Church, G. M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods 11, 499–507 (2014).
Reuter, J. A., Spacek, D. V. & Snyder, M. P. High-throughput sequencing technologies. Mol. Cell 58, 586–597 (2015).
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016).
Smithies, O., Gregg, R. G., Boggs, S. S., Koralewski, M. A. & Kucherlapati, R. S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317, 230–234 (1985).
DeWitt, M. A. et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl Med. 8, 360ra134 (2016).
Byrne, S. M., Ortiz, L., Mali, P., Aach, J. & Church, G. M. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 43, e21 (2015).
Dever, D. P. et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Boeke, J. D. et al. The Genome Project-Write. Science 353, 126–127 (2016).
Robertson, D. E. et al. A new dawn for industrial photosynthesis. Photosynth. Res. 107, 269–277 (2011).
Salhi, A. et al. DESM: portal for microbial knowledge exploration systems. Nucleic Acids Res. 44, D624–D633 (2016).
Fischer, C. R., Klein-Marcuschamer, D. & Stephanopoulos, G. Selection and optimization of microbial hosts for biofuels production. Metab. Eng. 10, 295–304 (2008).
Isaacs, F. J. et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 333, 348–353 (2011).
Lajoie, M. J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Wang, G. et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 20, 616–623 (2014).
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715 (2015).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015).
Matano, M. et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 (2015).
Schwank, G. et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658 (2013).
Liu, J. et al. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry 6, e703 (2016).
Soldner, F. et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99 (2016).
Chan, L. Y., Kosuri, S. & Endy, D. Refactoring bacteriophage T7. Mol. Syst. Biol. 1, 2005.0018 (2005).
Mandell, D. J. et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature 518, 55–60 (2015).
Rovner, A. J. et al. Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93 (2015).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Zhang, Y., Buchholz, F., Muyrers, J. P. & Stewart, A. F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20, 123–128 (1998).
Yu, D. et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA 97, 5978–5983 (2000).
Lacroix, B. & Citovsky, V. Transfer of DNA from bacteria to eukaryotes. mBio 7, e00863–16 (2016).
Xu, K., Stewart, A. F. & Porter, A. C. G. Stimulation of oligonucleotide-directed gene correction by Redβ expression and MSH2 depletion in human HT1080 cells. Mol. Cells 38, 33–39 (2015).
Rios, X. et al. Stable gene targeting in human cells using single-strand oligonucleotides with modified bases. PLoS ONE 7, e36697 (2012).
Igoucheva, O., Alexeev, V. & Yoon, K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 8, 391–399 (2001).
Aarts, M. & te Riele, H. Subtle gene modification in mouse ES cells: evidence for incorporation of unmodified oligonucleotides without induction of DNA damage. Nucleic Acids Res. 38, 6956–6967 (2010).
Quadros, R. M. et al. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18, 92 (2017).
Swarts, D. C. et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261 (2014).
Gao, F., Shen, X. Z., Jiang, F., Wu, Y. & Han, C. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 34, 768–773 (2016).
Lee, S. H. et al. Failure to detect DNA-guided genome editing using Natronobacterium gregoryi Argonaute. Nat. Biotechnol. 35, 17–18 (2016).
Burgess, S. et al. Questions about NgAgo. Protein Cell 7, 913–915 (2016).
Javidi-Parsijani, P. et al. No evidence of genome editing activity from Natronobacterium gregoryi Argonaute (NgAgo) in human cells. PLoS ONE 12, e0177444 (2017).
Kim, H. & Kim, J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334 (2014).
Chandrasegaran, S. & Carroll, D. Origins of programmable nucleases for genome engineering. J. Mol. Biol. 428, 963–989 (2016).
Chevalier, B. S. & Stoddard, B. L. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 29, 3757–3774 (2001).
Smith, J. et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 34, e149 (2006).
Chevalier, B. S. et al. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell 10, 895–905 (2002).
Boissel, S. et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 42, 2591–2601 (2014).
Wolfs, J. M. et al. MegaTevs: single-chain dual nucleases for efficient gene disruption. Nucleic Acids Res. 42, 8816–8829 (2014).
Birling, M.-C., Gofflot, F. & Warot, X. Site-specific recombinases for manipulation of the mouse genome. Methods Mol. Biol. 561, 245–263 (2009).
Sauer, B. & Henderson, N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl Acad. Sci. USA 85, 5166–5170 (1988).
Turan, S. & Bode, J. Site-specific recombinases: from tag-and-target-to tag-and-exchange-based genomic modifications. FASEB J. 25, 4088–4107 (2011).
Buchholz, F. & Stewart, A. F. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 19, 1047–1052 (2001).
Eroshenko, N. & Church, G. M. Mutants of Cre recombinase with improved accuracy. Nat. Commun. 4, 2509 (2013).
Santoro, S. W. & Schultz, P. G. Directed evolution of the site specificity of Cre recombinase. Proc. Natl Acad. Sci. USA 99, 4185–4190 (2002).
Saraf-Levy, T. et al. Site-specific recombination of asymmetric lox sites mediated by a heterotetrameric Cre recombinase complex. Bioorg. Med. Chem. 14, 3081–3089 (2006).
Gaj, T. et al. Enhancing the specificity of recombinase-mediated genome engineering through dimer interface redesign. J. Am. Chem. Soc. 136, 5047–5056 (2014).
Wallen, M. C., Gaj, T. & Barbas, C. F. Redesigning recombinase specificity for safe harbor sites in the human genome. PLoS ONE 10, e0139123 (2015).
Mastroianni, M. et al. Group II intron-based gene targeting reactions in eukaryotes. PLoS ONE 3, e3121 (2008).
Guo, H. et al. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science 289, 452–457 (2000).
Shalem, O., Sanjana, N. E. & Zhang, F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16, 299–311 (2015).
Tsai, S. Q. & Joung, J. K. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 17, 300–312 (2016).
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 (2016).
Yang, L. et al. Engineering and optimising deaminase fusions for genome editing. Nat. Commun. 7, 13330 (2016).
Fulco, C. P. et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016).
Canver, M. C. et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015).
Yang, L. et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350, 1101–1104 (2015).
Haimovich, A. D., Muir, P. & Isaacs, F. J. Genomes by design. Nat. Rev. Genet. 16, 501–516 (2015).
Smith, H. O., Hutchison, C. A., Pfannkoch, C. & Venter, J. C. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc. Natl Acad. Sci. USA 100, 15440–15445 (2003).
Gibson, D. G. et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220 (2008).
Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010).
Hutchison, C. A. et al. Design and synthesis of a minimal bacterial genome. Science 351, aad6253 (2016).
Lartigue, C. et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science 325, 1693–1696 (2009).
Ostrov, N. et al. Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822 (2016).
Dymond, J. S. et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 477, 471–476 (2011).
Annaluru, N. et al. Total synthesis of a functional designer eukaryotic chromosome. Science 344, 55–58 (2014).
Richardson, S. M. et al. Design of a synthetic yeast genome. Science 355, 1040–1044 (2017).
Shen, Y. et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science 355, eaaf4791 (2017).
Xie, Z.-X. et al. 'Perfect' designer chromosome V and behavior of a ring derivative. Science 355, eaaf4704 (2017).
Zhang, W. et al. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science 355, eaaf3981 (2017).
Mitchell, L. A. et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science 355, eaaf4831 (2017).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Tian, J. et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432, 1050–1054 (2004).
Gordon, D. et al. Long-read sequence assembly of the gorilla genome. Science 352, aae0344 (2016).
Loose, M., Malla, S. & Stout, M. Real-time selective sequencing using nanopore technology. Nat. Methods 13, 751–754 (2016).
Szalay, T. & Golovchenko, J. A. De novo sequencing and variant calling with nanopores using PoreSeq. Nat. Biotechnol. 33, 1087–1091 (2015).
Goodwin, S. et al. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 25, 1750–1756 (2015).
Fuller, C. W. et al. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc. Natl Acad. Sci. USA 113, 5233–5238 (2016).
Beliveau, B. J. et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147 (2015).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Frock, R. L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186 (2015).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Lensing, S. V. et al. DSBCapture: in situ capture and sequencing of DNA breaks. Nat. Methods 13, 855–857 (2016).
Osoegawa, K. et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 11, 483–496 (2001).
Straub, C., Granger, A. J., Saulnier, J. L. & Sabatini, B. L. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS ONE 9, e105584 (2014).
Agah, R. et al. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 100, 169–179 (1997).
Suzuki, K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016).
Pinder, J., Salsman, J. & Dellaire, G. Nuclear domain 'knock-in' screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 43, 9379–9392 (2015).
Chu, V. T. et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548 (2015).
Maruyama, T. et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33, 538–542 (2015).
Adamson, B., Smogorzewska, A., Sigoillot, F. D., King, R. W. & Elledge, S. J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 14, 318–328 (2012).
Agudelo, D. et al. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat. Methods 14, 615–620 (2017).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014).
He, X. et al. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 44, e85 (2016).
Maresca, M., Lin, V. G., Guo, N. & Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 23, 539–546 (2013).
Yang, Y. & Seed, B. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat. Biotechnol. 21, 447–451 (2003).
Bradley, A. et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm. Genome 23, 580–586 (2012).
Ordovás, L. et al. Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Rep. 5, 918–931 (2015).
Voziyanova, E. et al. Efficient Flp-Int HK022 dual RMCE in mammalian cells. Nucleic Acids Res. 41, e125 (2013).
Osterwalder, M. et al. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat. Methods 7, 893–895 (2010).
Zhang, X., Koolhaas, W. H. & Schnorrer, F. A versatile two-step CRISPR- and RMCE-based strategy for efficient genome engineering in Drosophila. G3 (Bethesda) 4, 2409–2418 (2014).
Bosley, K. S. et al. CRISPR germline engineering — the community speaks. Nat. Biotechnol. 33, 478–486 (2015).
Baltimore, D. et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science 348, 36–38 (2015).
Church, G. Perspective: encourage the innovators. Nature 528, S7 (2015).
Doudna, J. Perspective: embryo editing needs scrutiny. Nature 528, S6 (2015).
Schandera, J. & Mackey, T. K. Mitochondrial replacement techniques: divergence in global policy. Trends Genet. 32, 385–390 (2016).
Li, L. & Blankenstein, T. Generation of transgenic mice with megabase-sized human yeast artificial chromosomes by yeast spheroplast-embryonic stem cell fusion. Nat. Protoc. 8, 1567–1582 (2013).
Tomizuka, K. et al. Double trans-chromosomic mice: maintenance of two individual human chromosome fragments containing Ig heavy and kappa loci and expression of fully human antibodies. Proc. Natl Acad. Sci. USA 97, 722–727 (2000).
DiCarlo, J. E. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013).
EauClaire, S. F., Zhang, J., Rivera, C. G. & Huang, L. L. Combinatorial metabolic pathway assembly in the yeast genome with RNA-guided Cas9. J. Ind. Microbiol. Biotechnol. 43, 1001–1015 (2016).
Wang, S.-Z., Liu, B.-H., Tao, H. W., Xia, K. & Zhang, L. I. A genetic strategy for stochastic gene activation with regulated sparseness (STARS). PLoS ONE 4, e4200 (2009).
Gouble, A. et al. Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J. Gene Med. 8, 616–622 (2006).
Gellhaus, K., Cornu, T. I., Heilbronn, R. & Cathomen, T. Fate of recombinant adeno-associated viral vector genomes during DNA double-strand break-induced gene targeting in human cells. Hum. Gene Ther. 21, 543–553 (2010).
Robert, M.-A. et al. Efficacy and site-specificity of adenoviral vector integration mediated by the phage φC31 integrase. Hum. Gene Ther. Methods 23, 393–407 (2012).
Ou, H. et al. A highly efficient site-specific integration strategy using combination of homologous recombination and the ΦC31 integrase. J. Biotechnol. 167, 427–432 (2013).
Coluccio, A. et al. Targeted gene addition in human epithelial stem cells by zinc-finger nuclease-mediated homologous recombination. Mol. Ther. 21, 1695–1704 (2013).
Beaton, B. P. et al. Inclusion of homologous DNA in nuclease-mediated gene targeting facilitates a higher incidence of bi-allelically modified cells. Xenotransplantation 22, 379–390 (2015).
Nakade, S. et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 5, 5560 (2014).
Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L. & Corn, J. E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344 (2016).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Puchta, H., Dujon, B. & Hohn, B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21, 5034–5040 (1993).
Groth, A. C., Olivares, E. C., Thyagarajan, B. & Calos, M. P. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl Acad. Sci. USA 97, 5995–6000 (2000).
Bibikova, M. et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 21, 289–297 (2001).
Rouet, P., Smih, F. & Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA 91, 6064–6068 (1994).
Liu, P., Jenkins, N. A. & Copeland, N. G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476–484 (2003).
Testa, G. et al. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat. Biotechnol. 21, 443–447 (2003).
Epinat, J.-C. et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 31, 2952–2962 (2003).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Mercer, A. C., Gaj, T., Fuller, R. P. & Barbas, C. F. Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res. 40, 11163–11172 (2012).
Gaj, T., Mercer, A. C., Sirk, S. J., Smith, H. L. & Barbas, C. F. A comprehensive approach to zinc-finger recombinase customization enables genomic targeting in human cells. Nucleic Acids Res. 41, 3937–3946 (2013).
Owens, J. B. et al. Transcription activator like effector (TALE)-directed piggyBac transposition in human cells. Nucleic Acids Res. 41, 9197–9207 (2013).
Tsai, S. Q. et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32, 569–576 (2014).
Guilinger, J. P., Thompson, D. B. & Liu, D. R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 32, 577–582 (2014).
Pettersen, E. F. et al. UCSF Chimera — a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Zhang, J., McCabe, K. A. & Bell, C. E. Crystal structures of λ exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl Acad. Sci. USA. 108, 11872–11877 (2011).
Chen, Z., Yang, H. & Pavletich, N. P. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–484 (2008).
Sheng, G. et al. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl Acad. Sci. USA 111, 652–657 (2014).
Muñoz, I. G. et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 39, 729–743 (2011).
Gopaul, D. N., Guo, F. & Van Duyne, G. D. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 17, 4175–4187 (1998).
Peisach, E. & Pabo, C. O. Constraints for zinc finger linker design as inferred from X-ray crystal structure of tandem Zif268-DNA complexes. J. Mol. Biol. 330, 1–7 (2003).
Mak, A. N.-S., Bradley, P., Cernadas, R. A., Bogdanove, A. J. & Stoddard, B. L. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335, 716–719 (2012).
Toor, N., Keating, K. S., Taylor, S. D. & Pyle, A. M. Crystal structure of a self-spliced group II intron. Science 320, 77–82 (2008).
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014).
Hatada, S., Nikkuni, K., Bentley, S. A., Kirby, S. & Smithies, O. Gene correction in hematopoietic progenitor cells by homologous recombination. Proc. Natl Acad. Sci. USA 97, 13807–13811 (2000).
Wang, H. H. et al. Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593 (2012).
Liu, P.-Q. et al. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol. Bioeng. 106, 97–105 (2010).
Wang, J. et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 33, 1256–1263 (2015).
Hoban, M. D. et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125, 2597–2604 (2015).
Karakikes, I. et al. A comprehensive TALEN-based knockout library for generating human-induced pluripotent stem cell–based models for cardiovascular diseases. Circ. Res. 120, 1561–1571 (2017).
Donoho, G., Jasin, M. & Berg, P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol. Cell. Biol. 18, 4070–4078 (1998).
Lin, J. et al. Creating a monomeric endonuclease TALE-I-SceI with high specificity and low genotoxicity in human cells. Nucleic Acids Res. 43, 1112–1122 (2015).
Cai, D., Cohen, K. B., Luo, T., Lichtman, J. W. & Sanes, J. R. Improved tools for the Brainbow toolbox. Nat. Methods 10, 540–547 (2013).
Ding, Q. et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394 (2013).
Acknowledgements
This work was supported by NIH grant RM1HG008525 “Center for Genomically Engineered Organs”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
G.M.C. is a co-founder of Editas Medicine and eGenesis Bio and serves advisory roles in several companies involved in genome editing and engineering. A detailed listing of G.M.C.'s Tech Transfer, Advisory Roles and Funding Sources can be obtained from http://arep.med.harvard.edu/gmc/tech.html. R.C. declares no competing interests.
Related links
Glossary
- Homologous recombination
-
(HR). Exchange of sequence between two highly similar DNA molecules.
- Homology-directed repair
-
(HDR). The process by which double-strand breaks (DSBs) in DNA are repaired when in the presence of an additional DNA moiety that has homology to the DNA sequence surrounding the DSB.
- Protospacer adjacent motif
-
(PAM). A short, characteristic sequence (typically between 3 and 8 nucleotides in length) that the CRISPR-associated nuclease (such as Cas9 or Cpf1) recognizes before making its DNA break. Depending on the CRISPR system used, this sequence must either flank the 3′ end or 5′ end of the sequence of interest.
- Reading
-
The use of massively parallel sequencing technologies to decipher nucleotide composition.
- Writing
-
The use of either genome-editing tools and/or DNA synthesis to make desired changes in DNA sequence.
Rights and permissions
About this article
Cite this article
Chari, R., Church, G. Beyond editing to writing large genomes. Nat Rev Genet 18, 749–760 (2017). https://doi.org/10.1038/nrg.2017.59
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2017.59
This article is cited by
-
Genebanking plant genetic resources in the postgenomic era
Agriculture and Human Values (2023)
-
When is it Safe to Edit the Human Germline?
Science and Engineering Ethics (2021)
-
Efficient correction of a deleterious point mutation in primary horse fibroblasts with CRISPR-Cas9
Scientific Reports (2020)
-
Organizing genome engineering for the gigabase scale
Nature Communications (2020)