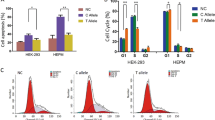
Abstract
Background
Non-syndromic orofacial cleft (NSOC) is one of the most common craniofacial malformations with complex etiology. This study aimed to explore the role of specific SNPs in ZFP36L2 and its functional relevance in zebrafish models.
Methods
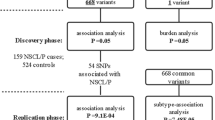
We analyzed genetic data of the Chinese Han population from two previous GWAS, comprising of 2512 cases and 2255 controls. Based on the Hardy-Weinberg Equilibrium (HWE) and minor allele frequency (MAF), SNPs in the ZFP36L2 were selected for association analysis. In addition, zebrafish models were used to clarify the in-situ expression pattern of zfp36l2 and the impact of its Morpholino-induced knockdown.
Results
Via association analysis, rs7933 in ZFP36L2 was significantly associated with various non-syndromic cleft lip-only subtypes, potentially conferring a protective effect. Zebrafish embryos showed elevated expression of zfp36l2 in the craniofacial region during critical stages of oral cavity formation. Furthermore, Morpholino-induced knockdown of zfp36l2 led to craniofacial abnormalities, including cleft lip, which was partially rescued by the addition of zfp36l2 mRNA.
Conclusion
Our findings highlight the significance of ZFP36L2 in the etiology of NSOC, supported by both human genetic association data and functional studies in zebrafish. These results pave the way for further exploration of targeted interventions for craniofacial malformations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
05 March 2024
A Correction to this paper has been published: https://doi.org/10.1038/s10038-024-01239-4
References
Smarius B, Loozen C, Manten W, Bekker M, Pistorius L, Breugem C. Accurate diagnosis of prenatal cleft lip/palate by understanding the embryology. World J Methodol. 2017;7:93–100.
Rahimov F, Jugessur A, Murray JC. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J. 2012;49:73–91.
Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–78.
Beaty TH, Hetmanski JB, Zeiger JS, Fan YT, Liang KY, VanderKolk CA, et al. Testing candidate genes for non-syndromic oral clefts using a case-parent trio design. Genet Epidemiol. 2002;22:1–11.
Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–7.
Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–6.
Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–9.
Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44:968–71.
Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun. 2015;6:6414.
Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet 2016;25:2862–72.
Yu Y, Zuo X, He M, Gao J, Fu Y, Qin C, et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 2017;8:14364.
Huang L, Jia Z, Shi Y, Du Q, Shi J, Wang Z, et al. Genetic factors define CPO and CLO subtypes of nonsyndromicorofacial cleft. PLoS Genet. 2019;15:e1008357.
Li B, Yong L, Yu Y, Yu Y, Zhen Q, Ge H, et al. Genome-wide analyses of nonsyndromic cleft lip with or without palate identify 20 new risk loci in the Chinese Han population. J Genet Genomics. 2022;49:903–5.
Leslie EJ, Liu H, Carlson JC, Shaffer JR, Feingold E, Wehby G, et al. A genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3. Am J Hum Genet. 2016;98:744–54.
Butali A, Mossey PA, Adeyemo WL, Eshete MA, Gowans LJJ, Busch TD, et al. Genomic analyses in African populations identify novel risk loci for cleft palate. Hum Mol Genet. 2019;28:1038–51.
He M, Zuo X, Liu H, Wang W, Zhang Y, Fu Y, et al. Genome-wide analyses identify a novel risk locus for nonsyndromic cleft palate. J Dent Res. 2020;99:1461–8.
Li MJ, Kumari P, Lin YS, Yao ML, Zhang BH, Yin B, et al. A variant in the IRF6 promoter associated with the risk for orofacial clefting. J Dent Res. 2023;102:806–13.
Sun JL, Shi JY, Yin B, Lin YS, Shi B, Jia ZL. Association analysis of SNPs in GRHL3, FAF1, and KCNJ2 with NSCPO sub-phenotypes in Han Chinese. Oral Dis. 2022;28:2204–14.
Zhang JX, Arneja JS. Evidence-based medicine: the bilateral cleft lip repair. Plast Reconstr Surg. 2017;140:152E–65E.
Carlson JC, Taub MA, Feingold E, Beaty TH, Murray JC, Marazita ML, et al. Identifying genetic sources of phenotypic heterogeneity in orofacial clefts by targeted sequencing. Birth Defects Res. 2017;109:1030–8.
Curtis SW, Chang D, Lee MK, Shaffer JR, Indencleef K, Epstein MP, et al. The PAX1 locus at 20p11 is a potential genetic modifier for bilateral cleft lip. HGG Adv. 2021;2:100025.
Yin B, Shi JY, Lin YS, Shi B, Jia ZL. SNPs at TP63 gene was specifically associated with right-side cleft lip in Han Chinese population. Oral Dis. 2021;27:559–66.
Zheng W, Sha QQ, Hu H, Meng F, Zhou Q, Chen X, et al. Biallelic variants in ZFP36L2 cause female infertility characterised by recurrent preimplantation embryo arrest. J Med Genet. 2022;59:850–7.
Xing R, Zhou Y, Yu J, Yu Y, Nie Y, Luo W, et al. Whole-genome sequencing reveals novel tandem-duplication hotspots and a prognostic mutational signature in gastric cancer. Nat Commun. 2019;10:2037.
Vogel KU, Bell LS, Galloway A, Ahlfors H, Turner M. The RNA-binding proteins Zfp36l1 and Zfp36l2 enforce the Thymic β-selection checkpoint by limiting DNA damage response signaling and cell cycle progression. J Immunol. 2016;197:2673–85.
Cook ME, Bradstreet TR, Webber AM, Kim J, Santeford A, Harris KM, et al. The ZFP36 family of RNA binding proteins regulates homeostatic and autoreactive T-cell responses. Sci Immunol. 2022;7:eabo0981.
Yonemori K, Seki N, Kurahara H, Osako Y, Idichi T, Arai T, et al. ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci. 2017;108:124–35.
Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–6.
Li MJ, Shi JY, Zhu QS, Shi B, Jia ZL. Targeted re-sequencing of the 2p21 Locus identifies non-syndromic cleft lip only novel susceptibility gene ZFP36L2. Front Genet. 2022;13:802229.
Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69.
Sun H, Li D, Chen S, Liu Y, Liao X, Deng W, et al. Zili inhibits transforming growth factor-beta signaling by interacting with Smad4. J Biol Chem. 2010;285:4243–50.
Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 2013;132:771–81.
Moreno Uribe LM, Fomina T, Munger RG, Romitti PA, Jenkins MM, Gjessing HK, et al. A population-based study of effects of genetic loci on orofacial clefts. J Dent Res. 2017;96:1322–9.
Pan Y, Han Y, Zhang H, Zhou L, Li D, Cai Q, et al. Association and cumulative effects of GWAS-identified genetic variants for nonsyndromic orofacial clefts in a Chinese population. Environ Mol Mutagen. 2013;54:261–7.
Deplancke B, Alpern D, Gardeux V. The genetics of transcription factor DNA binding variation. Cell. 2016;166:538–54.
Shen X, Song S, Li C, Zhang J. Synonymous mutations in representative yeast genes are mostly strongly non-neutral. Nature. 2022;606:725–31.
Einarsson H, Salvatore M, Vaagensø C, Alcaraz N, Bornholdt J, Rennie S, et al. Promoter sequence and architecture determine expression variability and confer robustness to genetic variants. Elife. 2022;11:e80943.
Rochard L, Monica SD, Ling IT, Kong Y, Roberson S, Harland R, et al. Roles of Wnt pathway genes wls, wnt9a, wnt5b, frzb and gpc4 in regulating convergent-extension during zebrafish palate morphogenesis. Development 2016;143:2541–7.
Yu Y, Alvarado R, Petty LE, Bohlender RJ, Shaw DM, Below JE, et al. Polygenic risk impacts PDGFRA mutation penetrance in non-syndromic cleft lip and palate. Hum Mol Genet. 2022;31:2348–57.
Maili L, Tandon B, Yuan Q, Menezes S, Chiu F, Hashmi SS, et al. Disruption of fos causes craniofacial anomalies in developing zebrafish. Front Cell Dev Biol. 2023;11:1141893.
Tréguer K, Faucheux C, Veschambre P, Fédou S, Thézé N, Thiébaud P. Comparative functional analysis of ZFP36 genes during Xenopus development. PLoS One. 2013;8:e54550.
Stumpo DJ, Broxmeyer HE, Ward T, Cooper S, Hangoc G, Chung YJ, et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114:2401–10.
Méndez-Maldonado K, Vega-López GA, Aybar MJ, Velasco I. Neurogenesis from neural crest cells: molecular mechanisms in the formation of cranial nerves and Ganglia. Front Cell Dev Biol. 2020;8:635.
Keyte A, Hutson MR. The neural crest in cardiac congenital anomalies. Differentiation. 2012;84:25–40.
Klaus A, Müller M, Schulz H, Saga Y, Martin JF, Birchmeier W. Wnt/β-catenin and Bmp signals control distinct sets of transcription factors in cardiac progenitor cells. Proc Natl Acad Sci USA. 2012;109:10921–6.
Graf D, Malik Z, Hayano S, Mishina Y. Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Factor Rev. 2016;27:129–39.
Manzari-Tavakoli A, Babajani A, Farjoo MH, Hajinasrollah M, Bahrami S, Niknejad H. The cross-talks among Bone Morphogenetic Protein (BMP) signaling and other prominent pathways involved in neural differentiation. Front Mol Neurosci. 2022;15:827275.
Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7:3.
Heijlen M, Houbrechts AM, Bagci E, Van Herck SL, Kersseboom S, Esguerra CV, et al. Knockdown of type 3 iodothyronine deiodinase severely perturbs both embryonic and early larval development in zebrafish. Endocrinology. 2014;155:1547–59.
Winata CL, Korzh S, Kondrychyn I, Zheng W, Korzh V, Gong Z. Development of zebrafish swimbladder: The requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev Biol. 2009;331:222–36.
Yin A, Korzh V, Gong Z. Perturbation of zebrafish swimbladder development by enhancing Wnt signaling in Wif1 morphants. Biochim Biophys Acta. 2012;1823:236–44.
Acknowledgements
The authors would like to thank everyone who contributed to this research. This project was supported by the National Natural Science Foundation of China (No. 82170919) and the Research and Develop Program, West China Hospital of Stomatology Sichuan University (No. RD-03-202301). The authors declare no potential conflicts of interest with respect to the authorship and/ or publication of this article.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: In this article Jialin Sun and Mujia Li should have been denoted as equally contributing authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, J., Li, M., Sun, H. et al. Genetic association and functional validation of ZFP36L2 in non-syndromic orofacial cleft subtypes. J Hum Genet 69, 139–144 (2024). https://doi.org/10.1038/s10038-024-01222-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-024-01222-z