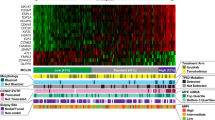
Abstract
Monitoring tumour burden and therapeutic response through analyses of circulating cell-free tumour DNA (ctDNA) and extracellular RNA (exRNA) in multiple myeloma (MM) patients were performed in a Phase Ib trial of 24 relapsed/refractory patients receiving oral azacitidine in combination with lenalidomide and dexamethasone. Mutational characterisation of paired BM and PL samples at study entry identified that patients with a higher number of mutations or a higher mutational fractional abundance in PL had significantly shorter overall survival (OS) (p = 0.005 and p = 0.018, respectively). A decrease in ctDNA levels at day 5 of cycle 1 of treatment (C1D5) correlated with superior progression-free survival (PFS) (p = 0.017). Evaluation of exRNA transcripts of candidate biomarkers indicated that high CRBN levels coupled with low levels of SPARC at baseline were associated with shorter OS (p = 0.000003). IKZF1 fold-change <0.05 at C1D5 was associated with shorter PFS (p = 0.0051) and OS (p = 0.0001). Furthermore, patients with high baseline CRBN coupled with low fold-change at C1D5 were at the highest risk of progression (p = 0.0001). In conclusion, this exploratory analysis has provided the first demonstration in MM of ctDNA for predicting disease outcome and of the utility of exRNA as a biomarker of therapeutic response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rasche L, Chavan SS, Stephens OW, Patel PH, Tytarenko R, Ashby C, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun. 2017;8:268.
Mithraprabhu S, Sirdesai S, Chen M, Khong T, Spencer A. Circulating tumour DNA analysis for tumour genome characterisation and monitoring disease burden in extramedullary multiple myeloma. Int J Mol Sci. 2018;19:1858. https://doi.org/10.3390/ijms19071858.
Mithraprabhu S, Khong T, Ramachandran M, Chow A, Klarica D, Mai L, et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia. 2017;31:1695–705.
Oberle A, Brandt A, Voigtlaender M, Thiele B, Radloff J, Schulenkorf A, et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica. 2017;102:1105–11.
Rustad EH, Coward E, Skytoen ER, Misund K, Holien T, Standal T, et al. Monitoring multiple myeloma by quantification of recurrent mutations in serum. Haematologica. 2017;102:1266–72.
Kis O, Kaedbey R, Chow S, Danesh A, Dowar M, Li T, et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun. 2017;8:15086.
Barzon L, Boscaro M, Pacenti M, Taccaliti A, Palu G. Evaluation of circulating thyroid-specific transcripts as markers of thyroid cancer relapse. Int J Cancer. 2004;110:914–20.
Chen XQ, Bonnefoi H, Pelte MF, Lyautey J, Lederrey C, Movarekhi S, et al. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6:3823–6.
Deligezer U, Yaman F, Darendeliler E, Dizdar Y, Holdenrieder S, Kovancilar M, et al. Post-treatment circulating plasma BMP6 mRNA and H3K27 methylation levels discriminate metastatic prostate cancer from localized disease. Clin Chim Acta. 2010;411:1452–6.
Garcia JM, Pena C, Garcia V, Dominguez G, Munoz C, Silva J, et al. Prognostic value of LISCH7 mRNA in plasma and tumor of colon cancer patients. Clin Cancer Res. 2007;13:6351–8.
Garcia V, Garcia JM, Pena C, Silva J, Dominguez G, Lorenzo Y, et al. Free circulating mRNA in plasma from breast cancer patients and clinical outcome. Cancer Lett. 2008;263:312–20.
Garcia V, Garcia JM, Silva J, Martin P, Pena C, Dominguez G, et al. Extracellular tumor-related mRNA in plasma of lymphoma patients and survival implications. PLoS ONE. 2009;4:e8173.
Joosse SA, Muller V, Steinbach B, Pantel K, Schwarzenbach H. Circulating cell-free cancer-testis MAGE-A RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with breast cancer and benign breast diseases. Br J Cancer. 2014;111:909–17.
Kang Y, Zhang J, Sun P, Shang J. Circulating cell-free human telomerase reverse transcriptase mRNA in plasma and its potential diagnostic and prognostic value for gastric cancer. Int J Clin Oncol. 2013;18:478–86.
Lo KW, Lo YM, Leung SF, Tsang YS, Chan LY, Johnson PJ, et al. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin Chem. 1999;45(8 Pt 1):1292–4.
March-Villalba JA, Martinez-Jabaloyas JM, Herrero MJ, Santamaria J, Alino SF, Dasi F. Cell-free circulating plasma hTERT mRNA is a useful marker for prostate cancer diagnosis and is associated with poor prognosis tumor characteristics. PLoS ONE. 2012;7:e43470.
Miura N, Maeda Y, Kanbe T, Yazama H, Takeda Y, Sato R, et al. Serum human telomerase reverse transcriptase messenger RNA as a novel tumor marker for hepatocellular carcinoma. Clin Cancer Res. 2005;11:3205–9.
Silva JM, Dominguez G, Silva J, Garcia JM, Sanchez A, Rodriguez O, et al. Detection of epithelial messenger RNA in the plasma of breast cancer patients is associated with poor prognosis tumor characteristics. Clin Cancer Res. 2001;7:2821–5.
Silva JM, Rodriguez R, Garcia JM, Munoz C, Silva J, Dominguez G, et al. Detection of epithelial tumour RNA in the plasma of colon cancer patients is associated with advanced stages and circulating tumour cells. Gut. 2002;50:530–4.
Sueoka E, Sueoka N, Iwanaga K, Sato A, Suga K, Hayashi S, et al. Detection of plasma hnRNP B1 mRNA, a new cancer biomarker, in lung cancer patients by quantitative real-time polymerase chain reaction. Lung Cancer. 2005;48:77–83.
Wong SC, Lo SF, Cheung MT, Ng KO, Tse CW, Lai BS, et al. Quantification of plasma beta-catenin mRNA in colorectal cancer and adenoma patients. Clin Cancer Res. 2004;10:1613–7.
Xu W, Zhou H, Qian H, Bu X, Chen D, Gu H, et al. Combination of circulating CXCR4 and Bmi-1 mRNA in plasma: A potential novel tumor marker for gastric cancer. Mol Med Rep. 2009;2:765–71.
Zhou D, Tang W, Liu X, An HX, Zhang Y. Clinical verification of plasma messenger RNA as novel noninvasive biomarker identified through bioinformatics analysis for lung cancer. Oncotarget. 2017;8:43978–89.
Kalff A, Khong T, Mithraprabhu S, Bergin K, Reynolds J, Bowen KM, et al. Oral azacitidine (CC-486) in combination with lenalidomide and dexamethasone in advanced, lenalidomide-refractory multiple myeloma (ROAR study). Leuk Lymphoma. 2019. (in press).
Cluzeau T, Robert G, Mounier N, Karsenti JM, Dufies M, Puissant A, et al. BCL2L10 is a predictive factor for resistance to azacitidine in MDS and AML patients. Oncotarget. 2012;3:490–501.
Kaiser MF, Johnson DC, Wu P, Walker BA, Brioli A, Mirabella F, et al. Global methylation analysis identifies prognostically important epigenetically inactivated tumor suppressor genes in multiple myeloma. Blood. 2013;122:219–26.
Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–5.
Locatelli SL, Papait R, Careddu G, Koschorke A, Stirparo GG, Balzarotti M, et al. Upregulation of cereblon expression by the DNA methyltransferase inhibitor azacytidine strongly enhances lenalidomide cytotoxicity in germinal center B-cell-like (GCB) and activated B-cell-like (ABC) diffuse large B-cell lymphoma (DLBCL). Blood. 2014;124:2253.
Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–35.
Eichner R, Heider M, Fernandez-Saiz V, van Bebber F, Garz AK, Lemeer S, et al. Immunomodulatory drugs disrupt the cereblon-CD147-MCT1 axis to exert antitumor activity and teratogenicity. Nat Med. 2016;22:735–43.
Kronke J, Kuchenbauer F, Kull M, Teleanu V, Bullinger L, Bunjes D, et al. IKZF1 expression is a prognostic marker in newly diagnosed standard-risk multiple myeloma treated with lenalidomide and intensive chemotherapy: a study of the German Myeloma Study Group (DSMM). Leukemia. 2017;31:1363–7.
An J, Ponthier CM, Sack R, Seebacher J, Stadler MB, Donovan KA, et al. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4(CRBN) ubiquitin ligase. Nat Commun. 2017;8:15398.
Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS. High-dimensional variable selection for survival data. J Am Stat Assoc. 2010;105:205–17.
Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J Clin Oncol. 2015;33:3911.
Broyl A, Kuiper R, van Duin M, van der Holt B, el Jarari L, Bertsch U, et al. High cereblon expression is associated with better survival in patients with newly diagnosed multiple myeloma treated with thalidomide maintenance. Blood. 2013;121:624–7.
Heintel D, Rocci A, Ludwig H, Bolomsky A, Caltagirone S, Schreder M, et al. High expression of cereblon (CRBN) is associated with improved clinical response in patients with multiple myeloma treated with lenalidomide and dexamethasone. Br J Haematol. 2013;161:695–700.
Schuster SR, Kortuem KM, Zhu YX, Braggio E, Shi CX, Bruins LA, et al. The clinical significance of cereblon expression in multiple myeloma. Leuk Res. 2014;38:23–8.
Kronke J, Knop S, Langer C. Prognostic impact of Ikaros expression in lenalidomide-treated multiple myeloma. Oncotarget. 2017;8:106163–4.
Sehgal K, Das R, Zhang L, Verma R, Deng Y, Kocoglu M, et al. Clinical and pharmacodynamic analysis of pomalidomide dosing strategies in myeloma: impact of immune activation and cereblon targets. Blood. 2015;125:4042–51.
Zhu YX, Braggio E, Shi CX, Kortuem KM, Bruins LA, Schmidt JE, et al. Identification of cereblon-binding proteins and relationship with response and survival after IMiDs in multiple myeloma. Blood. 2014;124:536–45.
Alcazar O, Achberger S, Aldrich W, Hu Z, Negrotto S, Saunthararajah Y, et al. Epigenetic regulation by decitabine of melanoma differentiation in vitro and in vivo. Int J Cancer. 2012;131:18–29.
Lopez-Girona A, Heintel D, Zhang LH, Mendy D, Gaidarova S, Brady H, et al. Lenalidomide downregulates the cell survival factor, interferon regulatory factor-4, providing a potential mechanistic link for predicting response. Br J Haematol. 2011;154:325–36.
Acknowledgements
We thank the staff and patients of Malignant Haematology & Stem Cell Transplantation, Alfred Hospital for the invaluable contribution towards sample collection.
Funding
Dr. Mithraprabhu was supported by a fellowship from the James and Elsie Borrowman Estate.
Author information
Authors and Affiliations
Contributions
SM and AS had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. SM and AS were involved in the study concept and design. SM, RM, TK, AK, KB, JH, IS, KB, MR, KW, BKLW, JR and AS contributed to acquisition, analysis and interpretation of data. SM, RM and AS drafted the manuscript. Critical revision of the manuscript was performed by all authors. Statistical analyses were performed by SM, RM and JR. Administrative, technical and/or material support were provided by TK, AK, KB, JH, IS, KB, MR, BKLW, KC and AS. The study was supervised by SM, TK, AK and AS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mithraprabhu, S., Morley, R., Khong, T. et al. Monitoring tumour burden and therapeutic response through analysis of circulating tumour DNA and extracellular RNA in multiple myeloma patients. Leukemia 33, 2022–2033 (2019). https://doi.org/10.1038/s41375-019-0469-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0469-x
This article is cited by
-
Liquid biopsy by analysis of circulating myeloma cells and cell-free nucleic acids: a novel noninvasive approach of disease evaluation in multiple myeloma
Biomarker Research (2023)
-
Circulating tumour DNA analysis predicts relapse and improves risk stratification in primary refractory multiple myeloma
Blood Cancer Journal (2023)
-
Multifaceted roles of extracellular RNAs in different diseases
Military Medical Research (2022)
-
Monitoring multiple myeloma in the peripheral blood based on cell-free DNA and circulating plasma cells
Annals of Hematology (2022)
-
Cell-free DNA for the detection of emerging treatment failure in relapsed/ refractory multiple myeloma
Leukemia (2022)