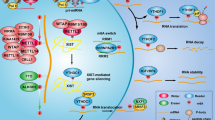
Abstract
Background
N6-methyladenosine (m6A) is the most abundant mRNA modification in mammals, participating in various biological processes. VIRMA is a key methyltransferase involved in m6A modification. However, the role of VIRMA in Hirschsprung’s disease (HSCR) remains unclear. This study aims to investigate the function of VIRMA in HSCR and identify its corresponding regulatory mechanisms.
Methods
The expression of VIRMA and GSK3β in colon tissues of HSCR was examined using RT-qPCR, Western blot, and Immunohistochemistry. Immunofluorescence detected localization of VIRMA and GSK3β. Cell proliferation was measured by CCK8 and EdU assays, and cell migration was evaluated via cell migration and wound healing assays. The stability of GSK3β mRNA was assessed using the actinomycin D assay and the overall level of m6A in cells was assessed by colorimetric assay.
Results
VIRMA was significantly downregulated in narrow-segment colon tissue. Silencing of VIRMA inhibited cell proliferation and migration. VIRMA can inhibit the degradation of GSK3β mRNA and increase the expression of GSK3β. GSK3β was significantly upregulated in narrow-segment colon tissues. Accordingly, our findings showed that GSK3β mediated the VIRMA-driven cell migration and proliferation.
Conclusion
VIRMA can inhibit cell migration and proliferation by upregulating the expression of GSK3β, contributing to the onset of HSCR.
Impact
-
The expressions of VIRMA were significantly reduced in HSCR, while GSK3β expression was increased in HSCR, and can be used as a molecular marker.
-
VIRMA overexpression promoted the proliferation and migration of SH-SY5Y and HEK-293T cells.
-
VIRMA can inhibit the degradation of GSK3β mRNA and increase the expression of GSK3β.
This is a preview of subscription content, access via your institution
Access options






Similar content being viewed by others
Data availability
Data included in this manuscript are available upon request by contacting the corresponding author and will be freely available to any researcher wishing to use them for non-commercial purposes without breaching participant confidentiality.
References
Jaroy, E. G. et al. “Too much guts and not enough brains”: (epi)genetic mechanisms and future therapies of Hirschsprung disease—a review. Clin. Epigenet. 11, 135 (2019).
Li, S. et al. Update on the pathogenesis of the hirschsprung-associated enterocolitis. Int. J. Mol. Sci. 24, 4602 (2023).
Jiang, Q. et al. RET somatic mutations are underrecognized in Hirschsprung disease. Genet. Med. 20, 770–777 (2017).
Morimoto, N. et al. Homozygous EDNRB mutation in a patient with Waardenburg syndrome type 1. Auris Nasus Larynx 45, 222–226 (2017).
Di Lascio, S. et al. Structural and functional differences in PHOX2B frameshift mutations underlie isolated or syndromic congenital central hypoventilation syndrome. Hum. Mutat. 39, 219–236 (2017).
Pingault, V., Zerad, L., Bertani-Torres, W. & Bondurand, N. SOX10: 20 years of phenotypic plurality and current understanding of its developmental function. J. Med. Genet. 59, 105–114 (2021).
Gao, Z. G. et al. Preliminary identification of key miRNAs, signaling pathways, and genes associated with Hirschsprung’s disease by analysis of tissue microRNA expression profiles. World J. Pediatr. 13, 489–495 (2017).
Villalba-Benito, L. et al. Overexpression of DNMT3b target genes during Enteric Nervous System development contribute to the onset of Hirschsprung disease. Sci. Rep. 7, 6221 (2017).
Huang, Z. et al. The m6A methyltransferase METTL3 affects cell proliferation and migration by regulating YAP expression in Hirschsprung disease. Pediatr. Surg. Int 39, 126 (2023).
Wang, B. et al. m6A demethylase ALKBH5 suppresses proliferation and migration of enteric neural crest cells by regulating TAGLN in Hirschsprung’s disease. Life Sci. 278, 119577 (2021).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Yue, Y., Liu, J. & He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Gene Dev. 29, 1343–1355 (2015).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Yu, J. et al. The m6 A readers YTHDF1 and YTHDF2 synergistically control cerebellar parallel fiber growth by regulating local translation of the key Wnt5a signaling components in axons. Adv. Sci. (Weinh.) 8, e2101329 (2021).
Wang, Y. et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206 (2018).
Wen, J. et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038.e6 (2018).
Yue, Y. et al. VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10 (2018).
Lan, T. et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 18, 186 (2019).
Miao, R. et al. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J. Cell Physiol. 235, 7420–7432 (2020).
Li, Y. et al. N6-methyladenosine methyltransferase KIAA1429 elevates colorectal cancer aerobic glycolysis via HK2-dependent manner. Bioengineered 13, 11923–11932 (2022).
Zhang, C. et al. Gene amplification-driven RNA methyltransferase KIAA1429 promotes tumorigenesis by regulating BTG2 via m6A-YTHDF2-dependent in lung adenocarcinoma. Cancer Commun. (Lond.) 42, 609–626 (2022).
Mancinelli, R. et al. Multifaceted roles of GSK-3 in cancer and autophagy-related diseases. Oxid. Med. Cell Longev. 2017, 4629495 (2017).
Kim, W. Y. et al. GSK-3 is a master regulator of neural progenitor homeostasis. Nat. Neurosci. 12, 1390–1397 (2009).
Morgan-Smith, M. et al. GSK-3 signaling in developing cortical neurons is essential for radial migration and dendritic orientation. Elife 3, e02663 (2014).
Butler Tjaden, N. E. & Trainor, P. A. The developmental etiology and pathogenesis of Hirschsprung disease. Transl. Res. 162, 1–15 (2013).
Torroglosa, A. et al. Epigenetic mechanisms in Hirschsprung disease. Int. J. Mol. Sci. 20, 3123 (2019).
Wang, C. X. et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 16, e2004880 (2018).
Chen, J. et al. m6A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genom. Proteom. Bioinf. 17, 154–168 (2019).
Qi, S. T. et al. N6-methyladenosine sequencing highlights the involvement of mRNA methylation in oocyte meiotic maturation and embryo development by regulating translation in Xenopus laevis. J. Biol. Chem. 291, 23020–23026 (2016).
Hu, Y. et al. Oocyte competence is maintained by m6A methyltransferase KIAA1429-mediated RNA metabolism during mouse follicular development. Cell Death Differ. 27, 2468–2483 (2020).
Jaworski, T., Banach-Kasper, E. & Gralec, K. GSK-3β at the intersection of neuronal plasticity and neurodegeneration. Neural Plast. 2019, 4209475 (2019).
Hu, S. et al. GSK3 inhibitors show benefits in an Alzheimer’s disease (AD) model of neurodegeneration but adverse effects in control animals. Neurobiol. Dis. 33, 193–206 (2008).
Dunning, C. J. et al. Direct high affinity interaction between Aβ42 and GSK3α stimulates hyperphosphorylation of tau. a new molecular link in Alzheimer’s disease? ACS Chem. Neurosci. 7, 161–170 (2015).
Funding
This study was supported by the Guangdong Basic and Applied Basic Research Foundation of China (2019A1515011086).
Author information
Authors and Affiliations
Contributions
Yang Yang, Mengzhen Zhang, Nan Li: preparation of the manuscript, statistics, and writing the manuscript. Chen Wang, Huirong Yang, Xinwei Hou: literature search, literature analyses. Jiaming Yang, Kaisi Fan: literature analyses and reviewed the manuscript. Kai Wu, Liucheng Yang: guide the project and revise the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the Ethics Committee of Zhujiang Hospital of Southern Medical University. Written informed consent was obtained from patients’ parents for the publication of any potentially identifiable images or data included in this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, Y., Zhang, M., Li, N. et al. Hirschsprung’s disease: m6A methylase VIRMA suppresses cell migration and proliferation by regulating GSK3β. Pediatr Res (2024). https://doi.org/10.1038/s41390-024-03136-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41390-024-03136-0