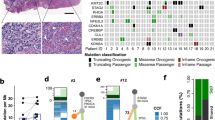
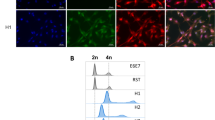
Abstract
MSCs (mesenchymal stem cells), responsible for tissue repair, rarely undergo cell fusion with somatic cells. Here, we show that ~5% of bladder cancer cells (UMUC-3) fuses with bone marrow-derived MSC (BM-MSC) in co-culture and maintains high tumorigenicity. In eleven fusion cell clones that have been established, Mb-scale deletions carried by the bladder cancer cells are mostly absent in the fusion cells, but copy number gains contributed by the cancer cells have stayed. Fusion cells exhibit increased populations of mitotic cells with 3-polar spindles, indicative of genomic instability. They grow faster in vitro and exhibit higher colony formation in anchorage-independent growth assay in soft agar than the parent UMUC-3 does. Fusion cells develop tumors, after 4 weeks of time lag, as efficiently as the parent UMUC-3 does in xenograft experiments. 264 genes are identified whose expression is specifically altered in the fusion cells. Many of them are interferon-stimulated genes (ISG), but are activated in a manner independent of interferon. Among them, we show that PD-L1 is induced in fusion cells, and its knockout decreases tumorigenesis in a xenograft model. PD-L1 is induced in a manner independent of STAT1 known to regulate PD-L1 expression, but is regulated by histone modification, and is likely to inhibit phagocytosis by PD1-expressing macrophages, thus protecting cancer cells from immunological attacks. The fusion cells overexpress multiple cytokines including CCL2 that cause tumor progression by converting infiltrating macrophages to tumor-associated-macrophage (TAM). The results present mechanisms of how cell fusion promotes tumorigenesis, revealing a novel link between cell fusion and PD-L1, and underscore the efficacy of cancer immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the finding of this study are openly available in figshare at https://doi.org/10.6084/m9.figshare.20140976 and in bioRxiv at https://doi.org/10.1101/2022.06.14.496068.
References
Hernández JM, Podbilewicz B. The hallmarks of cell-cell fusion. Development. 2017;144:4481–95.
Zhou X, Merchak K, Lee W, Grande JP, Cascalho M, Platt JL. Cell fusion connects oncogenesis with tumor evolution. Am J Pathol. 2015;185:2049–60.
Oren-Suissa M, Podbilewicz B. Cell fusion during development. Trends Cell Biol. 2007;17:537–46.
Weiler J, Dittmar T. Cell fusion in human cancer: the dark matter hypothesis. Cells. 2019;8:132. https://doi.org/10.3390/cells8020132.
Gast CE, Silk AD, Zarour L, Riegler L, Burkhart JG, Gustafson KT, et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv. 2018;4:eaat7828.
Yin L, Hu P, Shi X, Qian W, Zhau HE, Pandol SJ, et al. Cancer cell’s neuroendocrine feature can be acquired through cell-cell fusion during cancer-neural stem cell interaction. Sci Rep. 2020;10:1216.
Zhang LN, Zhang DD, Yang L, Gu YX, Zuo QP, Wang HY, et al. Roles of cell fusion between mesenchymal stromal/stem cells and malignant cells in tumor growth and metastasis. FEBS J. 2021;288:1447–56.
Pittenger MF, Discher DE, Péault BM, Phinney DG, Hare JM, Caplan AI. Mesenchymal stem cell perspective: cell biology to clinical progress. npj Regenerative Med. 2019;4:22.
Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63.
Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM, Fike JR, Lee HO, Pfeffer K, et al. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature. 2003;425:968–73.
Nygren JM, Jovinge S, Breitbach M, Säwén P, Röll W, Hescheler J, et al. Bone marrow-derived hematopoietic cells generate cardiomyocytes at a low frequency through cell fusion, but not transdifferentiation. Nat Med. 2004;10:494–501.
Rizvi AZ, Swain JR, Davies PS, Bailey AS, Decker AD, Willenbring H, et al. Bone marrow-derived cells fuse with normal and transformed intestinal stem cells. Proc Natl Acad Sci USA. 2006;103:6321–5.
Pawelek JM, Chakraborty AK. Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat Rev Cancer. 2008;8:377–86.
Johansson CB, Youssef S, Koleckar K, Holbrook C, Doyonnas R, Corbel SY, et al. Extensive fusion of haematopoietic cells with Purkinje neurons in response to chronic inflammation. Nat Cell Biol. 2008;10:575–83.
Rappa G, Mercapide J, Lorico A. Spontaneous formation of tumorigenic hybrids between breast cancer and multipotent stromal cells is a source of tumor heterogeneity. Am J Pathol. 2012;180:2504–15.
Nygren JM, Liuba K, Breitbach M, Stott S, Thorén L, Roell W, et al. Myeloid and lymphoid contribution to non-haematopoietic lineages through irradiation-induced heterotypic cell fusion. Nat Cell Biol. 2008;10:584–92.
Delespaul L, Merle C, Lesluyes T, Lagarde P, Le Guellec S, Perot G, et al. Fusion-mediated chromosomal instability promotes aneuploidy patterns that resemble human tumors. Oncogene. 2019;38:6083–94.
Feliciano D, Ott CM, Espinosa-Medina I, Weigel AV, Benedetti L, Milano KM, et al. YAP1 nuclear efflux and transcriptional reprograming follow membrane diminution upon VSV-G-induced cell fusion. Nat Commun. 2021;12:4502.
Ku JWK, Chen Y, Lim BJW, Gasser S, Crasta KC, Gan YH. Bacterial-induced cell fusion is a danger signal triggering cGAS-STING pathway via micronuclei formation. Proc Natl Acad Sci USA. 2020;117:15923–34.
Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–43.
Schoggins JW. Interferon-Stimulated Genes: What Do They All Do? Annu Rev Virol. 2019;6:567–84.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. (Methodological). 1995;57:289–300.
Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7.
Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini FC, Krause DS, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–7.
Tajima Y, Saito S, Ohno K, Tsukimura T, Tsujino S, Sakuraba H. Biochemical and structural study on a S529V mutant acid alpha-glucosidase responsive to pharmacological chaperones. J Hum Genet. 2011;56:440–6.
Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4.
Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–54.
Baker MJ, Goldstein AM, Gordon PL, Harbaugh KS, Mackley HB, Glantz MJ, et al. An interstitial deletion within 9p21.3 and extending beyond CDKN2A predisposes to melanoma, neural system tumours and possible haematological malignancies. J Med Genet. 2016;53:721–7.
Frigerio S, Disciglio V, Manoukian S, Peissel B, Della Torre G, Maurichi A, et al. A large de novo9p21.3 deletion in a girl affected by astrocytoma and multiple melanoma. BMC Med Genet. 2014;15:59.
Tanaka M, Grossman HB. In vivo gene therapy of human bladder cancer with PTEN suppresses tumor growth, downregulates phosphorylated Akt, and increases sensitivity to doxorubicin. Gene Ther. 2003;10:1636–42.
Aguirre-Ghiso JA, Sosa MS. Emerging topics on disseminated cancer cell dormancy and the paradigm of metastasis. Annu Rev Cancer Biol. 2018;2:377–93.
Li X, Yao W, Yuan Y, Chen P, Li B, Li J, et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–67.
BÜHring H-J, Battula VL, Treml S, Schewe B, Kanz L, Vogel W. Novel markers for the prospective isolation of human MSC. Ann N Y Acad Sci. 2007;1106:262–71. https://doi.org/10.1196/annals.1392.000
Miwa H, Era T. Tracing the destiny of mesenchymal stem cells from embryo to adult bone marrow and white adipose tissue via Pdgfrα expression. Development. 2018;145:dev155879. https://doi.org/10.1242/dev.155879.
Soliman H, Theret M, Scott W, Hill L, Underhill TM, Hinz B, et al. Multipotent stromal cells: One name, multiple identities. Cell Stem Cell. 2021;28:1690–707.
Miyamoto H, Yang Z, Chen Y-T, Ishiguro H, Uemura H, Kubota Y, et al. Promotion of bladder cancer development and progression by androgen receptor signals. JNCI: J Natl Cancer Inst. 2007;99:558–68.
Burr ML, Sparbier CE, Chan Y-C, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–5.
Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–10.
Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D–CDK4 kinase destabilizes PD-L1 via cullin 3–SPOP to control cancer immune surveillance. Nature. 2018;553:91–95.
Li C-W, Lim S-O, Xia W, Lee H-H, Chan L-C, Kuo C-W, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632.
Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2019;29:3766.
Kwak G, Kim D, Nam GH, Wang SY, Kim IS, Kim SH, et al. Programmed cell death protein ligand-1 silencing with polyethylenimine-dermatan sulfate complex for dual inhibition of melanoma growth. ACS Nano. 2017;11:10135–46.
Pelleitier M, Montplaisir S. The nude mouse: a model of deficient T-cell function. Methods Achiev Exp Pathol. 1975;7:149–66.
Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–9.
Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–5.
He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384–96.
Sun C, Mezzadra R, Schumacher TN. Regulation and Function of the PD-L1 Checkpoint. Immunity. 2018;48:434–52.
Li CW, Lim SO, Chung EM, Kim YS, Park AH, Yao J, et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 2018;33:187–201.e110.
Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95.
Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–39.
Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Investig. 2019;129:3324–38.
Sun LL, Yang RY, Li CW, Chen MK, Shao B, Hsu JM, et al. Inhibition of ATR downregulates PD-L1 and sensitizes tumor cells to T cell-mediated killing. Am J Cancer Res. 2018;8:1307–16.
Maniecki MB, Etzerodt A, Ulhøi BP, Steiniche T, Borre M, Dyrskjøt L, et al. Tumor-promoting macrophages induce the expression of the macrophage-specific receptor CD163 in malignant cells. Int J Cancer. 2012;131:2320–31.
Rubio C, Avendaño-Ortiz J, Ruiz-Palomares R, Karaivanova V, Alberquilla O, Sánchez-Domínguez R, et al. Toward tumor fight and tumor microenvironment remodeling: PBA induces cell cycle arrest and reduces tumor hybrid cells’ pluripotency in bladder cancer. Cancers. 2022;14:287.
Yu H, Liu T, Zhao Z, Chen Y, Zeng J, Liu S, et al. Mutations in 3′-long terminal repeat of HERV-W family in chromosome 7 upregulate syncytin-1 expression in urothelial cell carcinoma of the bladder through interacting with c-Myb. Oncogene. 2014;33:3947–58.
Acknowledgements
We thank Yoshinobu Iguchi and Kazunari Sekiyama for the preparation of cryosections. We also thank Yasumasa Nishito for assistance in the analyses with array-CGH, Daisuke Yamane for insightful discussion and comments. We thank Hiroyuki Sasanuma for critical reading of the manuscript. We thank the members of our laboratory for helpful discussion. This work was supported by JSPS KAKENHI Grant-in-Aid for Scientific Research (A) Grant Number 20H00463 (to HM) and by KAKENHI Grant-in-Aid for Scientific Research (C) Grant Number 18K07220 (to YT) and KAKENHI Grant-in-Aid for Challenging Exploratory Research Grant Number 16K15262 (to YT), respectively.
Author information
Authors and Affiliations
Contributions
Conceptualization: YT and HM; Methodology and Investigation: YT; Writing and Visualization: YT and HM; Supervision: HM and FS; Funding Acquisition and Project Administration: HM.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tajima, Y., Shibasaki, F. & Masai, H. Cell fusion upregulates PD-L1 expression for evasion from immunosurveillance. Cancer Gene Ther 31, 158–173 (2024). https://doi.org/10.1038/s41417-023-00693-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-023-00693-0