Abstract
Approximately 4.8–12.7 million tons of plastic waste has been estimated to be discharged into marine environments annually by wind and river currents. The Ellen MacArthur Foundation warns that the total weight of plastic waste in the oceans will exceed the total weight of fish in 2050 if the environmental runoff of plastic continues at the current rate. Hence, biodegradable plastics are attracting attention as a solution to the problems caused by plastic waste. Among biodegradable plastics, polyhydroxyalkanoates (PHAs) and poly(ε-caprolactone) (PCL) are particularly noteworthy because of their excellent marine biodegradability. In this review, the biosynthesis of PHA and cutin, a natural analog of PCL, and the biodegradation of PHA and PCL in carbon cycles in marine ecosystems are discussed. PHA is biosynthesized and biodegraded by various marine microbes in a wide range of marine environments, including coastal, shallow-water, and deep-sea environments. Marine cutin is biosynthesized by marine plants or obtained from terrestrial environments, and PCL and cutin are biodegraded by cutin hydrolytic enzyme-producing microbes in broad marine environments. Thus, biological carbon cycles for PHA and PCL exist in the marine environment, which would allow materials made of PHA and PCL to be quickly mineralized in marine environments.
Similar content being viewed by others
Marine environments are likely to be filled with plastics
Plastics are used in various aspects of our lives and are indispensable. In 2018, the global production of plastics reached 359 million ton [1]. Compared to when the industrial production of plastics began in 1950, there are currently ~200 times as many plastic products produced every year [2]. It has also been estimated that 300 million tons of plastic waste is generated annually, of which 79% is disposed of in landfills or discharged into the environment [3, 4]. In addition, 4.8–12.7 million tons of plastic waste, equivalent to 1.7–4.6% of that generated by coastal countries, is estimated to be discharged annually into the marine environment by wind and river currents [5]. Moreover, ~70% of the garbage at 37 ocean sampling points around the world have been found to be made up of plastics [6]. The Ellen MacArthur Foundation warns that the total weight of plastic waste in the oceans will exceed the total weight of fish in 2050 if the environmental runoff of plastic continues at this rate [7]. Large pieces of plastic debris spilled into the marine environment can be inadvertently ingested by or entangled with wildlife, causing their populations to decline and ultimately destroying the ecosystem. In particular, lost and found fishing gear (abandoned, lost, or otherwise discarded fishing gear: ALDFG), which is estimated to account for ~10% of marine plastic debris, continue to drift and entrap organisms in the ocean (ghost fishing), leading to significant economic problems [8]. In addition, persistent organic pollutants (POPs), which exist at low concentrations in the ocean, are concentrated on the surfaces of plastics discharged into the marine environment due to the hydrophobic properties of the plastic surface [9]. Plastic beads or pieces of plastic with a size of <5 mm spilled into the environment are called microplastics. There are concerns that if POPs present on microplastics are ingested by wildlife, they could be concentrated and stored in cells and tissues [10], eventually entering the human body through the food chain and resulting in adverse health effects [11].
Definition of biodegradable plastics
Biodegradable plastics are attracting attention as one of the solutions to the problems caused by plastic waste. Biodegradable plastics are materials degraded by the actions of microorganisms in the environment that are finally converted into inorganic substances. In general, the conversion rate of a material from an organic to an inorganic substance determines whether the material is a biodegradable plastic.
The degradation of biodegradable plastics proceeds in two stages (Fig. 1). The first stage is the degradation of the main chain of the polymer by microbial hydrolytic enzymes or nonbiological hydrolysis and the consequent formation of molecules of a size that can be taken up by microorganisms. The second stage is the mineralization of the decomposed products during catabolism, i.e., their conversion into carbon dioxide, nitrogen oxide, methane, and water [12]. Most of the international methods for evaluating the biodegradability of plastics are based on the quantification of carbon dioxide production or biochemical oxygen demand during the decomposition process [13].
Enzymatic and environmental degradability of biodegradable plastics
Table 1 [14,15,16,17,18,19] shows the properties of biodegradable plastics. Biodegradable plastics are obtained by microbial synthesis and chemical synthesis (Fig. 2).
Poly(3-hydroxyalkanoate) (PHA) is produced by microorganisms as an energy storage material. In contrast, poly(ε-caprolactone) (PCL), poly(ethylene succinate) (PESu), poly(butylene succinate) (PBSu), poly(butylene succinate-co-adipate) (PBSA), poly(butylene adipate-co-terephthalate) (PBAT), and poly(lactic acid) (PLA) are produced from petroleum or biomass via chemical synthesis. Biodegradable plastics are hydrolyzed by enzymes produced by microorganisms [20]. However, chemically synthesized biodegradable plastics do not have a dedicated degradation enzyme, unlike PHA [21]. Chemically synthesized biodegradable plastics are recognized and hydrolyzed in the environment as substrate analogs by lipases, which use fat as a substrate, and by cutinases, which uses the polyester cutin (present in the cuticular layer of plants), as a substrate [22,23,24,25,26,27,28,29] (Table 2).
Due to its structural similarity to polyalanine, PLA can also be degraded by some types of proteolytic enzymes, such as Proteinase K [30, 31] (Table 2).
Biodegradable plastics have different degradation rates in different environments, depending on their type (Table 3) [32,33,34,35,36,37,38,39].
With the exception of PHA and PCL, the biodegradation rate of such plastics in a marine environment is generally low. This means that many products currently available as biodegradable plastics cannot be readily degraded in marine environments. Therefore, there is a concern that they may damage the marine ecosystem in a manner similar to the damage caused by commodity plastics. In addition to the aliphatic polyesters PHA and PCL, several polymers, such as cellulose [40,41,42], agarose [43, 44], proteins [45], and poly(vinyl alcohol) (PVA) [46], have been suggested to exhibit relatively high biodegradability in marine environments.
This review focuses on two biodegradable polyesters, PHA and PCL, because they have clear carbon cycles in marine ecosystems and could be used as thermoplastics. The biosynthesis of PHA and cutin as a natural analog of PCL, as well as the biodegradation of PHA and PCL in carbon cycles, are described in a marine ecosystem.
What is PHA?
Some bacteria and archaea produce PHA, a type of aliphatic polyester, as an energy storage material when nutrient sources such as nitrogen and phosphate are suppressed due to the presence of excessive carbon sources in the environment [14]. In PHA-producing bacteria, PHA exists in an amorphous state as native PHA granules [47]. Once these granules are extracted from the cells with solvent, they are partially crystallized and used as a biodegradable thermoplastic. A variety of PHAs with different structures are produced depending on the type of bacterial species and carbon source provided [48]. Based on the number of carbons present in a monomer, PHA can be classified into three groups: short-chain PHA (≤5 carbons), medium-chain PHA (6–14 carbons), and long-chain PHA (more than 15 carbons). Among them, a homopolyester composed of R-3-hydroxybutanoic acid, known as poly(3-hydroxybutanoic acid) [P(3HB)], is the most common PHA produced by many microorganisms.
P(3HB) is a thermoplastic biodegradable polymer with a melting point of ~180 °C and thermal and mechanical properties similar to those of isotactic polypropylene (PP). P(3HB) is a hard and brittle material with high crystallinity and low elongation at break (~5%) [49]. Copolymerization with another hydroxyalkanoate (HA) unit as a second component can change the thermal and mechanical properties of P(3HB), as the second component can act as a defective factor in its crystal structure, thus reducing the degree of crystallinity [50,51,52]. For example, in the case of poly(3-hydroxybutanoic-co-17 mol% 3-hydroxyhexanoic acid) (PHBHHx), in which P(3HB) is copolymerized with 3-hydroxyhexanoic acid (3HHx) as the second component, the melting point of PHBHHx decreases from ~180 to 130 °C, and the crystallinity decreases from 60 + 5% to 29 + 5% compared to the P(3HB) homopolymer. At the same time, the tensile strength decreases from 43 to 20 MPa, and the elongation at break changes from 6 to 850% [51, 53]. Thus, P(3HB) can be changed from a hard and brittle material to a flexible material by copolymerization with a second component.
Mechanism of PHA biosynthesis
The synthesis of P(3HB) begins with the conversion of 2 molecules of acetyl-CoA to acetoacetyl-CoA catalyzed by β-ketothiolase (PhaA, EC 2.3.1.9). Next, NADPH-dependent acetoacetyl-CoA reductase (PhaB, EC 1.1.1.36) converts acetoacetyl-CoA into the P(3HB) monomer (R)-3-hydroxybutyryl-CoA (3HB-CoA), which is polymerized by PHB polymerase (PhaC, EC 2.3.1-), resulting in the formation of P(3HB) [54]. In addition, the intermediate produced in the fatty acid biosynthetic pathway, (R)-3-hydroxyacyl ACP ((R)-3HA-ACP), is converted to (R)-3HA-CoA by (R)-3HA-ACP transferase (PhaG, EC 2.4.1-) to provide a PHA monomer [55, 56]. In Aeromonas caviae, also known as a PHBHHx-synthesizing bacteria and a type of Pseudomonas species, (R)-specific enoyl-CoA hydratase (PhaJ, EC 4.2.1.119) hydrates enoyl-CoA, which is an intermediate in the β-oxidation pathway, to produce 3HA-CoA as a PHA monomer [57]. PHA synthase is classified into four classes (I, II, III, and IV). The class I PHA polymerase (EC 2.3.1.B2) from Cupriavidus necator, which shows specificity for (R)-3HA-CoA, a monomer having 3–5 carbons, consists of a homodimeric protein comprising a single subunit (PhaC) with a molecular weight of ~60 kDa [58]. The class II PHA polymerase (EC 2.3.1.B3) from Pseudomonas spp., which shows specificity for (R)-3HA-CoA with 6–14 carbons, is a homodimeric protein consisting of a single subunit (PhaC) with a molecular weight of ~60 kDa [59]. The class III PHA polymerase (EC 2.3.1.B4) from Allochromatium vinosum is a heterodimeric enzyme composed of a 40 kDa PhaC subunit and a PhaE subunit of ~40 kDa, which shows a specificity for (R)-3HA-CoA with 3–5 carbons. The class IV PHA polymerase present in Bacillus megaterium is a heterodimeric enzyme composed of a PhaC subunit of ~40 kDa and a PhaR subunit of ~20 kDa, which has specificity for (R)-3-hydroxyacyl-CoA with 3–5 carbons. However, other PHA monomeric components (3HHx, 3HO, 4HB, 6HHx) are also available as substrates [60, 61].
PHA-producing bacteria in marine environments
Since the discovery by Lemoigne in 1925 that B. megaterium stores P(3HB), several microbes have been found to synthesize PHA in a variety of environments [62]. As listed in Table 4, various species of PHA-synthesizing microorganisms have also been isolated from a wide range of marine environments—from shallow to deep seawater [63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98].
PHA-synthesizing bacteria belonging to the phyla Proteobacteria, Firmicutes, and Actinobacteria have been isolated from shallow water and surrounding marine sediments below a depth of 200 m. Burkholderia sp. AIU M5M02, isolated from shallow sea mud, can synthesize P(3HB) using mannitol from seaweed as a carbon source [72]. Sulfate-reducing bacteria (SRBs), such as the Desulfococcus–Desulfosarcina group, Desulfococcus multivorans DSM 2059 strain, Desulfobotulus sapovorans DSM 2055 strain, Desulfosarcina variabilis DSM 2060, Desulfonema magnum DSM 2077, and Desulfobacterium autotrophicum HRM2 strain, which have contributed to carbon and sulfur cycling in the marine environment, have synthesized P(3HB) and PHBV [75]. Halomonas hydrothermalis isolated from coastal waters synthesizes P(3HB) using crude glycerol, a Jatropha biodiesel byproduct, as a carbon source [78]. Massilia sp. UMI-21 isolated from seaweed synthesizes PHA using starch, maltotriose, and maltose as carbon sources [81]. Pseudomonas guezennei isolated from atoll microbial mats uses glucose as a single carbon source to synthesize a medium-chain 3HA copolymer consisting of 3HB, 3HHx, 3-hydroxyoctanoate (3HO), 3-hydroxydecanoate (3HD), 3-hydroxydodecenoate (3Hdde), and 3‐hydroxydodecanoate (3HDd) [88]. The Vibrio sp. KN01 strain isolated from seawater was found to synthesize P(3HB) using glucose, fructose, gluconic acid, and soybean oil as carbon sources [98]. Thus, PHA-producing bacteria that inhabit shallow waters can synthesize various forms of PHA from carbon sources such as polysaccharides, sugars, organic acids, fatty acids, and glycerol that are derived from either marine plants or from fats/oils and their degradation products.
An oxygen-evolving photosynthetic cyanobacteria, Spirulina subsalsa, has been isolated from shallow seawater as a PHA producer. This strain biosynthesized 5.9% P(3HB) per dry cell weight from carbon dioxide in a marine medium [94]. In addition, aerobic nonoxygenic photosynthetic bacteria (aerobic anoxygenic phototrophic bacteria: AAPB) are widely distributed in marine environments and are known to be PHA producers. Bacteria classified in the genera Dinoroseobacter, Roseobacter, Labrenzia, and Erythrobacter, which are marine AAPBs isolated from marine microalgae, synthesize PHA in the presence of sugars or organic acids. Among them, PHA production by Dinoroseobacter sp. JL1447 increased with light irradiation [77]. Three species of purple sulfur bacteria (Thiohalocapsa marina, Thiophaeococcus mangrovi, and Marichromatium bheemlicum) and nine species of red nonsulfur bacteria (purple nonsulfur bacteria) (Afifella marina, five species of Rhodovulum and three species of Roseospira) were isolated [91]. Likewise, various PHA producers have been found in marine organisms. Photobacterium phosphoreum is present in the luminescent organs and intestines of fish [86], and bacteria belonging to the genera Vibrio [99], Bacillus [67], and Brevibacterium [71] that cohabitate with sponges have the ability to synthesize PHA.
PHA-producing bacteria have also been isolated from the deep sea, organisms living in the deep sea, and deep seafloor soils, although the numbers of such bacteria are fewer than those found in the shallow sea. Colwellia sp. JAMM-0421 and Moritella sp. strain JCM21335, isolated from deep seawater, synthesize PHA from sugars and fats [74]. Pseudoalteromonas sp. SM9913 isolated from deep sea bottom soil synthesized copolymers of 3HD and 3HDD using either glucose, decanoic acid, or olive oil as a single carbon source [87]. Halomonas profundus AT1214, isolated from deep sea hydrothermal vent shrimp, synthesizes P(3HB) from sugars and organic acids [79].
The diversity of the PHA synthase gene, phaC, in seawater from seven sampling sites at a depth of 24–5373 m was assessed by phaC amplicon analysis. A PhaC amino acid sequence fragment encoded by the gene obtained from the marine amplicon analysis revealed that in marine environments, the known class I PhaC homologs from Chromobacterium, Marinobacter, Rhodospirillum, Acetobacteraceae bacterium, Oceanibaculum, and Oceanicaulis were the most dominant, followed by the class I PhaC homolog of betaproteobacteria and class I and class II PhaC homologs of gammaproteobacteria [100].
In addition to reports on microbial PHA synthesis in marine environments, marine metagenomic analysis thus suggests that PHA-synthesizing bacteria inhabit a wide range of marine environments, from shallow waters to deep waters, thereby contributing to the production of PHA in the ocean.
Degradation of PHA in marine environments
PHA is known to be highly biodegradable in various marine environments. Microbes that produce extracellular PHA-degrading enzymes [such as P(3HB) depolymerase (EC 3.1.1.75)] are widespread in the ocean. Enzymatic degradation products of PHA are metabolized and mineralized by marine microorganisms. PHA is also produced in the ocean, as mentioned above, and is also degraded and mineralized in the ocean. Therefore, PHA can be regarded as a polymer in which the carbon cycle is established not only in the terrestrial environment but also in the marine environment. Some studies on the biodegradation of PHA in the ocean are given below.
P(3HB), PHBV, and P(3HB-co-4HB) films were installed in a seawater circulation tank in the waters around Jogashima Island, Kanagawa, Japan; the thickness of the films was observed to decrease by 13–22 µm after 3 weeks, and the thickness of the PHBV films was observed to decrease by 100–140 µm after 17 weeks. After 8 weeks of environmental exposure, the weight of the PHBV film decreased by 65% compared to the initial weight, and the elongation at break and the stress at break were zero [101]. However, the molecular weight of the specimens did not change before or after exposure. A P(3HB) film was installed in Akabane Port, Aichi, Japan, and after one week, several craters had appeared. Two weeks later, the weight of the P(3HB) film was reduced by ~60% from the initial weight [102]. When a P(3HB) film was incubated at 25 °C in seawater collected from the Pacific Ocean in Aichi, Japan, 9% of the initial weight was lost after 10 weeks, but the molecular weight and crystallinity of the specimens did not change before and after incubation. Moreover, after 3 weeks of immersion in seawater, the tensile strength of P(3HB) decreased to ~60% of the initial value [103]. After 6 weeks of PHBV film immersion in Baltic Seawater, it was observed that the film weight decreased by 60% compared to the initial weight, and its surface became rougher [104]. When P(3HB) and PHBV films (73 ± 5 mg, 30 mm in diameter, 0.1 mm thick) and injection-molded products (330 ± 25 mg, 10 mm in diameter) were immersed in the South China Sea the weights of the films decreased by 42% and 46%, respectively, after 160 days compared to the initial weights, and the weights of injection-molded parts decreased by 38% and 13%, respectively [105]. After P(3HB), PHBV, and P(3HB-co-4HB) were added to seawater collected from Tokyo Bay and Oarai Beach, Ibaraki, Japan at 25 °C under aerobic conditions for 28 days, the weight of each film decreased by 23–41%, 100%, and 59–70%, respectively, compared to the initial weights. According to the MITI method, the BOD biodegradability of P(3HB), PHBV and P(3HB-co-4HB) reached 14–27%, 78–84%, and 43–51%, respectively, after incubation for 28 days at 25 °C in seawater [34]. When P(3HB) and PHBV were incubated at 30 °C in artificial seawater with 13 marine microbial inoculum sources in accordance with the active standard, ASTM D6691, all samples showed more than 70% degradation after incubation for 40 days. When P(3HB) and PHBV films were incubated in seawater and submarine soil collected from Woods Hole Harbor at 21 °C for 49 days, their weights reduced by ~90% compared to their initial weights. Moreover, after P(3HB) and PHBV films were incubated in a seawater circulation tank in Woods Hole Harbor (12–22 °C, pH 7.9–8.1) for 90 days, their weights decreased by 33–70% compared to their initial weight [106]. BOD biodegradation testing of P(3HB) and its copolymers of seawater samples suggests that there is a positive correlation between biodegradability and temperatures in the range of 10–27 °C [32]. The degradability of PHBV in seawater with submarine soil collected from Lorient harbor in the Atlantic Ocean, France was 90% after 210 days [107].
PHBV fibers were immersed in deep seawater collected from Rausu, Toyama, and Kume, Japan for 12 months, and their tensile strength was measured every 3 months. On the one hand, the tensile strength of PHBV fibers immersed in seawater collected from Kume was completely lost after 3 months. On the other hand, after 3 months of immersion in seawater collected from Toyama, ~80% of the initial tensile strength of PHBV fibers was retained, and ~30% was retained after 12 months. Holes and cracks were found on the surface of the PHBV fibers after immersion in seawater [36]. Thus, PHA was shown to be biodegradable in a wide range of marine environments, including coastal, shallow-water, and deep-sea environments.
Environmental exposure tests of PHBV at four coastal sites in Puerto Rico suggested that the period when PHBV degradation begins is positively correlated to both viable cell counts and PHBV-degrading microbial counts [108]. However, there was no significant difference in the ratio of P(3HB)-degrading microbial counts to viable cell counts in soil, freshwater, or seawater [soil: 0.003–33% (median 4.4, average 9.3); freshwater: 0.59–25% (median 15, average 13); and seawater: 0.24–43.8% (median 2.5, average 9.7)]. The viable cell counts decreased in the following order: soil [1.3 × 105–1.6 × 108 c.f.u./g (median 5.3 × 106, average 1.1 × 107)], freshwater [2.3 × 102–3.2 × 105 c.f.u./g (median 9.6 × 103, average 6.9 × 104)], and seawater [1.6 × 102–3.1 × 105 c.f.u./g (median 7.9 × 102, average 5.4 × 103)] [109]. The time required for 1 mm thick PHBV films to be fully biodegraded was generally shorter in soil and freshwater environments [anaerobic sludge (6 weeks), estuarine sediment (40 weeks), activated sludge (60 weeks), and soil (75 weeks)] and was longer in marine environments [seawater (350 weeks)] [33]. Considering these data, it is highly likely that both microbial density and the total number of degrading microbes in the environment determine the lag time for the initiation of PHA degradation in the environment and the subsequent rate of degradation. Based on previously reported data regarding PHA degradation in shallow-water marine environments, coastal areas, and estuaries, Dilkes-Hoffma et al. estimated that the average degradation rate of PHA would be 0.04–0.09 mg/day/cm2. Employing the estimated degradation rate, they predicted that it would take 1.5–3.5 years for a 800 µm thick PHBV bottle to be fully biodegraded [110].
Recent studies have revealed that the surfaces of plastic materials spilled into natural environments create an ecosystem that is completely different than that of natural environments. The microbiota that form on such a plastic surface has been designated a “plastisphere” by the marine ecologist Zettler et al. [111]. Recently, it has become clear that the plastisphere plays an important role in the degradation of biodegradable plastics in marine environments. When PHBHHx was aerobically incubated at 23 °C in seawater collected from Takasago Port, Japan, a plastisphere was found to be formed on the surface, which contained PHA-degrading bacteria [112]. Shotgun metagenomic analysis of microflora formed on PHA, PET, and ceramic surfaces showed that the abundance of the P(3HB) depolymerase protein included in the plastisphere on the PHA surface was more than 20 times higher than those on PET, ceramic surfaces, or seawater [113]. Thus, it is presumed that when PHA is exposed to the marine environment, PHA-degrading microbial species are enriched in the plastispheres, and the enzymes produced by these microbes degrade PHA to compounds with a low molecular mass, followed by mineralization by the surrounding microorganisms.
Taken together, the environmental degradation of PHA is influenced by interrelated factors, such as biological factors (microbiota structure, abundance of degrading microbes, biofilm formation capacity, etc.), environmental factors (location, temperature, nutrients, sunlight and UV light, dissolved oxygen, salinity, pH, etc.), and characteristic features of the polymers (primary structure, shape, crystallinity, thermal properties, etc.).
PHA-degrading bacteria in marine environments
As described in the previous section, the biodegradation of PHA has been demonstrated in a wide range of marine environments. PHA-degrading bacteria have also been isolated from various marine environments in which PHA is degraded (Table 5) [112, 114,115,116,117,118,119,120,121,122,123,124,125].
Hence, the high biodegradability of PHA in a particular marine environment can be correlated with the ubiquity of its degrading microorganisms.
Three PHA-degrading bacteria, classified in the phylum gammaproteobacteria and isolated from marine environments [Pseudomonas stutzeri YM1006 (coastal seawater, Jogashima, Japan) [123], Shewanella sp. JKCM-AJ-6,1α (coastal seawater, Yaizu Port, Japan) [124], and Marinobacter sp. NK-1 (deep seawater, Sagami Bay, Japan) [120]] have been studied in detail. These strains grew well in 0.5 M NaCl, which is similar to seawater. When P. stutzeri YM1006 was cultured in the presence of P(3HB) and (R)-3HB, it secreted and produced P(3HB) depolymerase in the culture supernatant. Although it grew well in the presence of (S)-3HB, this enzyme was not produced. Shewanella sp. JKCM-AJ-6,1α produced P(3HB) depolymerase in the presence of P(3HB) and (R)-3HB; however, it did not produce this enzyme in the presence of (S)-3HB. Marinobacter sp. NK-1 strains grew well using several organic acids and amino acids, in addition to P(3HB), (R)-3HB, and (S)-3HB. This strain secreted P(3HB) depolymerase in the presence of P(3HB) and (S)-3HB. Streptomyces sp. SNG9 isolated from marine sediment expressed only weak P(3HB) depolymerase activity in the copresence of easy-to-use carbon sources, in addition to P(3HB). These data suggest that P(3HB) and its degradation products are likely candidates that act as inducers of the production of P(3HB) depolymerase, while easy-to-use carbon sources may repress its production [125].
Structure and function of marine P(3HB) depolymerases
Extracellular P(3HB) depolymerases (EC 3.1.1.75) are a group of enzymes that hydrolyze PHA composed of short-chain length hydroxyalkanoic acids (C3-C5): poly(3-hydroxypropionate) [P(3HP)], P(3HB), poly(3-hydroxyvalerate) (P(3HV)), and poly(4-hydroxybutyrate) [P(4HB)]. Although some of the enzymes that have the same EC numbers have hydrolytic activity against amorphous PHA in cells [intracellular P(3HB) depolymerases], they are different from extracellular P(3HB) depolymerases. Here, we describe extracellular P(3HB) depolymerases that hydrolyze crystalline PHA and are associated with the environmental degradation of plastics composed of PHAs.
Since P(3HB), the substrate for extracellular P(3HB) depolymerases, is a solid substance, the enzymatic reaction of P(3HB) depolymerase is a heterogeneous system that occurs at the solid–liquid interface. For efficient enzymatic reactions in such a heterogeneous system, P(3HB) depolymerases consist of a catalytic domain (CD) containing an active site, a substrate binding domain (SBD) for binding enzyme molecules to the surface of P(3HB), and a linker domain (LD) connecting these two functional domains. This multidomain structure (CD+SBD) is also commonly found in insoluble polymer hydrolytic enzymes such as cellulase and chitinase. The CDs of P(3HB) depolymerases contain three catalytic residues (Ser-His-Asp), of which the serine residue is the active center that cleaves the ester bond. The CDs of the enzyme are classified as serine hydrolases belonging to the α/β hydrolase superfamily [23].
Homologs of P(3HB) depolymerase genes have been found in the genomic DNA of various marine bacteria. To date, several P(3HB) depolymerase proteins from marine bacteria have been characterized (Table 6a, b) [114, 115, 119, 120, 123, 124, 126,127,128].
The CDs of P(3HB) depolymerases are classified into types A and B based on the position of the lipase box (Gly-X-Ser-X-Gly) contained within the CD. Both of these types are in a circular permutation relationship with each other, and both are presumed to have higher order structures that are made up of α/β hydrolase folds [129]. All of the marine P(3HB) depolymerases characterized to date are only of type A, in which the lipase box is located at the center (Fig. 3).
Schematic representative of marine and terrestrial P(3HB) depolymerases. CD catalytic domain, LD linker domain, SBD substrate binding domain. Fn(III), Cad, Thr indicate fibronectin type III-, cadherin-like-, and threonine-rich linker domains, respectively. In type A, the lipase box is located at the center of the CD, while in type B, it is located near the amino terminus of the CD
There are three types of LDs in P(3HB) depolymerases: fibronectin type III [Fn(III)], and cadherin (Cad), and threonine (Thr) types. Among them, Fn(III)- and Cad-type sequences were found in the LD of marine P(3HB) depolymerases. As an exception, the P(3HB) depolymerase from Bacillus sp. NRRL B-14911 consists of two types of LDs, LD1 and LD2, both of which did not show homology with any of the known LDs [119].
P(3HB) depolymerases of terrestrial microbes are composed of one CD, one LD, and one SBD, whereas all marine P(3HB) depolymerases have two SBDs in addition to one CD and one LD. Therefore, the molecular masses of marine P(3HB) depolymerases (60–70 kDa) are larger than those of terrestrial enzymes (40–50 kDa) [21, 128, 130]. Compared with the average substrate binding constant between the P(3HB) surface and the SBD of terrestrial P(3HB) depolymerase (K = 1.0 ml/µg) [21], those of marine P(3HB) depolymerases were as small as 0.2 ml/µg. In other words, this feature of the domain structure of marine P(3HB) depolymerases (i.e., having two SBDs) may compensate for its weak binding. Although the P(3HB) depolymerase produced by the marine bacterium Shewanella sp. JKCM-AJ-6,1α encodes two SBDs (SBDI and SBDII) at the locus, both were proteolytically cleaved during the secretion and production of the enzyme. The substrate binding constant of this enzyme that lacks two SBDs was the lowest among all the marine P(3HB) depolymerases (0.18 ± 0.01 ml/µg, Table 6b). This low binding constant is presumed to be complemented by a relative increase in hydrophobic interactions caused by a decrease in the dielectric constant due to the action of salt in seawater [131]. Thus, some characteristics of marine P(3HB) depolymerases differ from their terrestrial counterparts.
Marine P(3HB) depolymerases produced by marine species, such as Alcaligenes faecalis AE122, Shewanella sp. JKCM-AJ-6, 1α, Marinobacter sp. NK-1, and P. stutzeri YM1006 were characterized (Table 6b). Except for the enzyme from Shewanella sp. JKCM-AJ-6, 1α, those from the other species showed thermostability in the same temperature range as the sea temperature (−2 to 35 °C) [132]. On the other hand, the P(3HB) depolymerase from Shewanella sp. JKCM-AJ-6, 1α showed similar thermolability to most of the general enzymes active in marine environments. All marine P(3HB) depolymerases are stable in the marine pH range of 7.8–8.2 [133]. The P(3HB) depolymerase from Shewanella sp. JKCM-AJ-6, 1α was found to have no enzymatic activity at low salinity; however, the activity of the enzyme was enhanced at a NaCl concentration of 0.5 M, which is similar to that of seawater. By contrast, the addition of 0.5 M NaCl did not alter the activity of P(3HB) depolymerase from Marinobacter sp. NK-1. These results indicate that these P(3HB) depolymerases have an effect on the hydrolytic activity of P(3HB) in the marine environment.
What is PCL?
PCL is a linear aliphatic polyester composed of 6-hydroxyhexanoic acid as its constituent, which can be chemically synthesized by the ring-opening polymerization of ε-caprolactone. The glass transition temperature and melting points of PCL are −65 to −60 and 56–65 °C, respectively [15]. PCL is used as an impact-modifying additive for plastics, a ceramic binder, a binder for metal molds, a hot-melt adhesive, a heat-transfer ink, a flow modifier for resin molding, and a refractive index modifier. Although PCL is a chemically produced synthetic polymer, unlike PHA, PCL is known to be biodegradable in a variety of environments because it is recognized by microorganisms as an analog of the natural polymeric compound cutin [24].
The carbon cycle of cutin in the ocean
The cuticular layer on the surface of plants (mosses, gymnosperms, and angiosperms) functions as an extracellular hydrophobic layer to protect these plants from external environmental stresses and desiccation [134]. The cuticular layer contains mainly C16 and C18 hydroxy fatty acid condensed polyesters, known as cutins. Typical cutin monomers are composed of 10,16-dihydroxyhexadecanoic acid, 16-hydroxyhexadecanoic acid, 18-hydroxy-9,10-epoxyoctadecanoic acid, 9,10,18-trihydroxyoctadecanoic acid, and 18-hydroxyoctadec-9-enoic acid; the composition of which depends on the plant species. These monomers are linked to each other and to glycerol by an ester bond, resulting in cutin consisting of a straight chain and branched domain structure (Fig. 4a). Cutin biosynthesis in plants starts with de novo C16 or C18 fatty acid synthesis in the plastid of epidermal cells. Fatty acids are transported to the endoplasmic reticulum, where acyl-CoA is synthesized by the catalytic activity of the long-chain fatty acid CoA ligase (EC 6.2.1.3). ω-Hydroxylase, midchain hydroxylase, and epoxidase belonging to the cytochrome P450 family 77 subfamily A (EC 1.14.-) oxidize the ends and midchain carbons of cutin monomer-CoA, which is then converted into monoacylglycerol after its transfer to glycerol 3-phosphate by acyltransferase (EC 2.3.1.-). Next, monoacylglycerol is transported to the cuticular layer via the ABC transporter. In the cuticular layer, monoacylglycerol is polymerized by GDSL-motif lipase/hydrolase (EC 3.1.1.3-) to form cutin [135, 136].
a Cutin domain structure and b comparison of the chemical structures of cutin monomers with the PCL trimer [24]
Cutin also exists in the marine environment. Marine cutin has two main origins. One is associated with the influx of terrestrial environmental plants into the ocean, and the other is derived from marine plants. C16 and C18 hydroxy fatty acids, characteristic of terrestrial higher plants, have been detected in seabed sediments, providing evidence for the influx of cutin from a terrestrial environment to the sea [137,138,139,140]. The cutin content of marine sediments was observed to decrease from areas near an estuary to the deep sea to the seafloor below 1000 m, where 99.4% was found to be lost. This suggests that terrestrial cutin entering the ocean is almost completely degraded in the ocean [137]. The leaf surfaces of marine plants, such as Thalassia testudinum, Posidonia australis, and Zostera marina L., also have a cuticular layer, which contains cutin as a constituent in its interior [141,142,143,144].
Through this mechanism, a carbon cycle is formed in which cutin is biosynthesized in marine environments or transported from terrestrial environments and subsequently biodegraded in the marine environment.
Marine degradability of PCL
PCL, a biodegradable plastic, exhibits high biodegradability in a variety of environments. It is known to show high biodegradability similar to that exhibited by PHA in marine environments. The reason for this is presumably due to the structural similarity of PCL to cutin, where PCL enters the carbon cycle of cutin and is biodegraded in the ocean.
Exposure of PCL specimens to water collected from Bohai Bay for 52 weeks resulted in numerous depressions on the surface of the specimens and a decrease in their mechanical strength. The weights of the specimens were reduced by 29.8% compared to the initial weight [145]. After the PCL specimens were immersed in water obtained from Akabane Port, Aichi, Japan for 5 weeks, the tensile strength was completely lost, the elongation at break was 0%, the Young’s modulus was reduced to ~50% of the initial value, and the weight was reduced by ~34% of the initial weight [102]. When PCL was incubated in a buffer containing seawater collected from Tokyo Bay and the Pacific Ocean for 28 days at 25 °C under aerobic conditions, PCL reached a BOD biodegradation of 56–79% [34]. When PCL was incubated in a buffer containing seawater collected from Osaka Bay for 17 days at 27 °C under aerobic conditions, PCL showed a BOD biodegradation of 14.5–40.9% [32]. After 12 months of PCL specimens exposure to deep seawater drawn from Rausu, Toyama, and Kume, many pores and cracks were found on the surface of the specimens, and their tensile strength was completely lost [36].
PCL-degrading bacteria in the marine environment
PCL shows good degradability in various marine environments, and PCL-degrading microorganisms have been found in a wide range of marine environments. Bacteria belonging to the genus Pseudomonas isolated from marine environments, such as P. litoralis, P. oceani, and P. pelagia, demonstrated the ability to degrade PCL [146]. A PCL-degrading bacterium closely related to Pseudomonas pachastrellae was isolated from a plastic debris surface in a coastal environment. The strain grew well under NaCl concentrations similar to those of seawater (specific growth rate at 0.5 M NaCl: 0.3 h−1), suggesting that this strain is a typical marine bacterium. In the presence of PCL and its degradation product, 6-hydroxyhexanoic acid, PCL-degrading enzymatic activity was expressed in the culture supernatant by this strain in a marine medium [147]. Streptomyces sp. SM14 isolated from marine sponges degraded PCL [148]. Marine species of the genus Alcanivorax, including Alcanivorax sp. 24, A. xenomutans JC109, and A. dieselolei B-5 formed clear zones on PCL-containing plates [114]. Shewanella sp., Moritella sp., Psychrobacter sp., and Pseudomonas sp. were isolated as PCL-degrading bacteria from deep sea sediments of the Kurile and Japan Trenches. Among them, Shewanella sp. CT01 and Moritella spp. CT12 and JT01 are psychrophilic and piezophilic bacteria adapted to deep sea environments [149].
PCL-degrading enzymes: cutinases and lipases
Cutinases (E.C.3.1.1.74) are enzymes that hydrolyze the carboxy ester bonds of cutins, while lipases (E.C. 3.1.1.3) are enzymes that hydrolyze triacylglycerols. Both enzymes belong to the α/β hydrolase superfamily and contain three catalytic residues (Ser-His-Asp) and two amino acid residues that form an oxyanion hole to stabilize the transition state of the substrate. Nucleophilic attack by the hydroxyl group of the catalytic serine on the carbon of the substrate carbonyl group forms an acyl intermediate, and nucleophilic attack of this intermediate by water molecules results in hydrolysis of the ester bond [150,151,152]. Cutinases and lipases tend to have an affinity for short-chain ester substrates and long-chain ester substrates, respectively. However, it is difficult to classify both of these enzymes on the basis of substrate specificity alone, since there are cutinases that degrade long-chain triacylglycerols and lipases that degrade cutin [153, 154]. It is also known that both enzymes differ in their structure around the active site. There are two types of lipases, one that retains the lid domain covering the active site and the other that does not [150]. However, almost all cutinases do not have a lid domain [152]. Binding of the micelle surface of a fat substrate to the lid domain changes its structure and increases the area of the hydrophobic surface around the active site, thus increasing the interaction between the substrate and active site. This phenomenon is referred to as “interfacial activation” and helps to increase the enzymatic activity at the liquid–liquid interface of oil and water. By contrast, the interfacial activation effect of the lid domain is small at the solid–liquid interface, such as polyester and water, while the lid domain can physically inhibit the hydrolytic activity of an enzyme at the solid–liquid interface [155]. The effect of the presence or absence of the lid domain on the hydrolysis of polyesters was investigated using lipase from Rhizopus oryzae and cutinase from Fusarium solani, and it was found that easy access to the active site of the enzyme due to the absence of the lid domain enhanced the rate of polyester hydrolysis [156]. The lipases from Bacillus pumilus [29] and Moesziomyces antarcticus (homotypic synonym: Candida antarctica [157, 158]), which have PCL hydrolysis activity, lack a lid domain. These findings suggest that lipases and cutinases that do not retain the lid domain may be involved in the hydrolysis of PCL and other polyester substrates.
Marine PCL hydrolytic enzyme candidates
The α/β-hydrolase (CDS: ABO2449) of Alcanivorax borkumensis, which was expressed in Escherichia coli, was found to have the ability to hydrolyze a variety of aliphatic polyesters, such as PLA, PBSA, and PESu in addition to PCL. The gene for this enzyme is homologous (identity level 42–90%) to genes encoding putative proteins of hydrocarbon-degrading microorganisms, including Alcanivorax spp., Hoyosella subflava (homotypic synonym: Amycolicicoccus subflavus), Marinobacter nanhaiticus, and Hydrocarboniphaga effusa. This enzyme displayed high specificity for a short acyl chain of C2–C4. It was active in the pH range of 7–11 and in the temperature range of 4–70 °C and was also active in the presence of 3 M NaCl. Considering that the pH of the marine environment is 7.8–8.2 [133] and the water temperature ranges from −2 to 35 °C [132], this enzyme may be involved in PCL hydrolysis in the marine environment [159]. A PCL-degrading enzyme, Esterase SM14est, which is produced by the marine bacterium Streptomyces sp. SM14, possesses a conserved lipase box (Gly-X1-Ser-X2-Gly) and catalytic triad. This enzyme has 9 β-sheet and 7 α-helix structures and belongs to the α/β- hydrolase superfamily [148].
Compared to studies on cutinases or lipases with PCL-degrading activity from terrestrial microorganisms, there are very few studies related to marine microorganisms. Nevertheless, recent marine genomic analysis suggests that PCL-degrading enzymes are generally found in marine environments. For example, 349 homologs of cutinase-like enzyme genes were detected in 31 marine metagenomes and 11 terrestrial metagenomes from 108 marine water samples and 25 terrestrial samples. An enzyme with PET hydrolytic activity, PET 2, which was cloned from a marine metagenomics data set, could degrade PCL [160]. Cutinase-like enzyme (PE-H) produced by the marine bacterium Pseudomonas aestusnigri VGXO, which possesses PCL-hydrolytic activity, exhibited 48–51% homology with polyester hydrolases from the terrestrial species Streptomyces viridis, Thermobifida sp., and Ideonella sakaiensis [161]. These findings indicate that cutinase-like enzymes, which catalyze PCL hydrolysis, are present in the marine environment as well as in the terrestrial environment.
Induction of PCL degradation activity by PCL
The expression of cutinases produced by Fusarium vanettenii (homotypic synonym: F. solani f. sp. pisi) and F. verticillioides (heterotypic synonym: F. moniliforme) require the hydrolysates of PCL, which function as inducers. This could be due to structural homology with a trimer of PCL and the cutin constituent, 10,16-dihydroxy C16 acid, in the structure of 9,10,18-trihydroxy C18 acid [24] (Fig. 4b). Although the cutinase of Aspergillus oryzae degrades PBSu, the constituents of PBSu, succinate, and 1,4-butanediol induce the enzyme [25].
A marine bacterium, Pseudomonas sp. TKCM64, isolated from marine debris, not only exhibited PCL-hydrolytic activity but also had PESu hydrolytic activity. In the presence of PCL, its degradation products or cutin monomers, this strain expressed both PCL and PESu degradation activity. However, this strain showed relatively low polyester degradation activity in the presence of succinic acid and ethylene glycol, which are components of PESu. From these data, it can be inferred that chemically synthesized polyesters other than PCL and their constituents do not function as inducers of the said degradative enzymes in this strain [147]. This may provide insight into the low marine degradability of chemically synthesized polyesters other than PCL.
Conclusion
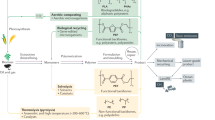
To date, biodegradable plastics that degrade in soil and compost have been developed, but most of them show negligible degradation in the ocean. Among them, the biodegradable polyesters PHA and PCL showed good biodegradability in marine environments where biological carbon cycles exist for both polyesters. This allows the materials made of PHA and PCL to be taken up into these carbon cycles and quickly mineralized. Consequently, such materials showed good biodegradability in marine environments. The marine environment has a low density of microorganisms compared to other environments, which makes it difficult to rapidly degrade biodegradable plastics. However, from the viewpoint of utilizing the biological carbon cycles inherently present in the ocean, it is possible to sufficiently increase the biodegradation rate of biodegradable plastics in the ocean (Figs. 5 and 6). A great deal of information is still needed to understand the biological carbon cycle in the ocean if we aim to develop plastics that are fully biodegrade at a sufficient rate in the ocean. We hope that this review provides a starting point for more research on this subject in the future.
References
Plastics Europe. Plastics—the Facts 2019; 2019. https://www.plasticseurope.org/application/files/9715/7129/9584/FINAL_web_version_Plastics_the_facts2019_14102019.pdf.
Ritchie H, Roser M. Plastic pollution. Our World in Data; 2018. https://ourworldindata.org/plastic-pollution.
UNEP. Single-use plastics: a roadmap for sustainability; 2018. https://wedocs.unep.org/bitstream/handle/20.500.11822/25496/singleUsePlastic_sustainability.pdf?sequence=1&isAllowed=y.
Geyer R, Jambeck JR, Law KL. Production, use, and fate of all plastics ever made. Sci Adv. 2017;3:e1700782. https://doi.org/10.1126/sciadv.1700782.
Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, et al. Plastic waste inputs from land into the ocean. Science. 2015;347:768–71. https://doi.org/10.1126/science.1260352.
Derraik JGB. The pollution of the marine environment by plastic debris: a review. Mar Pollut Bull. 2002;44:842–52.
World Economic Forum. Ellen MacArthur Foundation. The new plastics economy: rethinking the future of plastics; 2016. www3.weforum.org/docs/WEF_The_New_Plastics_Economy.pdf.
Stelfox M, Hudgins J, Sweet M. A review of ghost gear entanglement amongst marine mammals, reptiles and elasmobranchs. Mar Pollut Bull. 2016;11:6–17.
Rios LM, Moore C, Jones PR. Persistent organic pollutants carried by synthetic polymers in the ocean environment. Mar Pollut Bull. 2007;54:1230–7.
Rochman CM, Hoh E, Kurobe T, Teh SJ. Ingested plastic transfers hazardous chemicals to fish and induces hepatic stress. Sci Rep. 2013;3:32632. https://doi.org/10.1038/srep03263.
Setala O, Fleming-Lehtinen V, Lehtiniemi M. Ingestion and transfer of microplastics in the planktonic food web. Environ Pollut. 2014;185:77–83.
Lucas N, Bienaime C, Belloy C, Queneudec M, Silvestre F, Nava-Saucedo JE. Polymer biodegradation: mechanisms and estimation techniques. Chemosphere. 2008;73:429–42.
Harrison JP, Boardman C, O’Callaghan K, Delort AM, Song J. Biodegradability standards for carrier bags and plastic films in aquatic environments: a critical review. R Soc Open Sci. 2018;5:171792. https://doi.org/10.1098/rsos.171792.
Sudesh K, Abe H, Doi Y. Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog Polym Sci. 2000;25:1503–55.
Labet M, Thielemans W. Synthesis of polycaprolactone: a review. Chem Soc Rev. 2009;38:3484–504.
Tserki V, Matzinos P, Pavlidou E, Vachliotis D, Panayiotou C. Biodegradable aliphatic polyesters. Part I. Properties and biodegradation of poly(butylene succinate-co-butylene adipate). Polym Degrad Stabil. 2006;91:367–76.
Yamamoto M, Witt U, Skupin G, Beimborn D, Müller J. Biodegradable aliphatic-aromatic polyesters: “Ecoflex®”. Biopolymers Online; 2005.
Nampoothiri KM, Nair NR, John RP. An overview of the recent developments in polylactide (PLA) research. Bioresour Technol. 2010;101:8493–501.
Ku H, Wang H, Pattarachaiyakoop N, Trada M. A review on the tensile properties of natural fiber reinforced polymer composites. Compos B Eng. 2011;42:856–73.
Jendrossek D, Schirmer A, Schlegel HG. Biodegradation polyhydroxyalkanoic acids. Appl Microbiol Biot. 1996;46:451–63.
Kasuya K, Ohura T, Masuda K, Doi Y. Substrate and binding specificities of bacterial polyhydroxybutyrate depolymerases. Int J Biol Macromol. 1999;24:329–36.
Tokiwa Y, Suzuki T. Hydrolysis of polyesters by lipases. Nature. 1977;270:76–78.
Jaeger KE, Steinbuchel A, Jendrossek D. Substrate specificities of bacterial polyhydroxyalkanoate depolymerases and lipases: bacterial lipases hydrolyze poly(omega-hydroxyalkanoates). Appl Environ Microbiol. 1995;61:3113–8.
Murphy CA, Cameron JA, Huang SJ, Vinopal RT. Fusarium polycaprolactone depolymerase is cutinase. Appl Environ Microbiol. 1996;62:456–60.
Maeda H, Yamagata Y, Abe K, Hasegawa F, Machida M, Ishioka R, et al. Purification and characterization of a biodegradable plastic-degrading enzyme from Aspergillus oryzae. Appl Microbiol Biotechnol. 2005;67:778–88.
Murphy CA, Cameron JA, Huang SJ, Vinopal RT. A second polycaprolactone depolymerase from Fusarium, a lipase distinct from cutinase. Appl Microbiol Biotechnol. 1998;50:692–6.
Ando Y, Yoshikawa K, Yoshikawa T, Nishioka M, Ishioka R, Yakabe Y. Biodegradability of poly(tetramethylene succinate-co-tetramethylene adipate): I. Enzymatic hydrolysis. Polym Degrad Stabil. 1998;61:129–37.
Shinozaki Y, Morita T, Cao X, Yoshida S, Koitabashi M, Watanabe T, et al. Biodegradable plastic-degrading enzyme from Pseudozyma antarctica: cloning, sequencing, and characterization. Appl Microbiol Biotechnol. 2013;97:2951–9.
Muroi F, Tachibana Y, Soulenthone P, Yamamoto K, Mizuno T, Sakurai T, et al. Characterization of a poly(butylene adipate-co-terephthalate) hydrolase from the aerobic mesophilic bacterium Bacillus pumilus. Polym Degrad Stabil. 2017;137:11–22.
Tokiwa Y, Calabia BP. Biodegradability and biodegradation of poly(lactide). Appl Microbiol Biotechnol. 2006;72:244–51.
Williams DF. Enzymic hydrolysis of polylactic acid. Eng Med. 1981;10:5–7.
Nakayama A, Yamano N, Kawasaki N. Biodegradation in seawater of aliphatic polyesters. Polym Degrad Stabil. 2019;166:290–9.
Luzier WD. Materials derived from biomass biodegradable materials. Proc Natl Acad Sci USA. 1992;89:839–42.
Kasuya K, Takagi K, Ishiwatari S, Yoshida Y, Doi Y. Biodegradabilities of various aliphatic polyesters in natural waters. Polym Degrad Stabil. 1998;59:327–32.
Teramoto N, Urata K, Ozawa K, Shibata M. Biodegradation of aliphatic polyester composites reinforced by abaca fiber. Polym Degrad Stabil. 2004;86:401–9.
Sekiguchi T, Saika A, Nomura K, Watanabe T, Fujimoto Y, Enoki M, et al. Biodegradation of aliphatic polyesters soaked in deep seawaters and isolation of poly(ε-caprolactone)-degrading bacteria. Polym Degrad Stabil. 2011;96:1397–403.
Fujimaki T. Processability and properties of aliphatic polyesters, ‘BIONOLLE’, synthesized by polycondensation reaction. Polym Degrad Stabil. 1998;59:209–14.
Witt U, Müller RJ, Deckwer WD. New biodegradable polyester-copolymers from commodity chemicals with favorable use properties. J Environ Polym Degrad. 1995;3:215–23.
Bagheri AR, Laforsch C, Greiner A, Agarwal S. Fate of so‐called biodegradable polymers in seawater and freshwater. Glob Chall. 2017;1:1700048. https://doi.org/10.1002/gch2.201700048.
Zambrano MC, Pawlak JJ, Daystar J, Ankeny M, Goller CC, Venditti RA. Aerobic biodegradation in freshwater and marine environments of textile microfibers generated in clothes laundering: effects of cellulose and for polyester-based microfibers on the microbiome. Mar Pollut Bull. 2020;151:110826. https://doi.org/10.1016/j.marpolbul.2019.110826.
Taylor LE, Henrissat B, Coutinho PM, Ekborg NA, Hutcheson SW, Weiner RA. Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40T. J Bacteriol. 2006;188:3849–61.
Yang JC, Madupu R, Durkin SA, Ekborg NA, Pedamallu CS, Hostetler JB, et al. The Complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (shipworms). PLoS ONE. 2009;4:e6085. https://doi.org/10.1371/journal.pone.0006085.
Sawant SS, Salunke BK, Taylor LE, Kim BS. Enhanced agarose and xylan degradation for production of polyhydroxyalkanoates by co-culture of marine bacterium, Saccharophagus degradans and its contaminant, Bacillus cereus. Appl Sci. 2017;7. https://doi.org/10.3390/app7030225.
Liu G, Wu SM, Jin WH, Sun CM. Amy63, a novel type of marine bacterial multifunctional enzyme possessing amylase, agarase and carrageenase activities. Sci Rep. 2016;6:18726. https://doi.org/10.1038/srep18726.
Ramesh S, Mathivanan N. Screening of marine actinomycetes isolated from the Bay of Bengal, India for antimicrobial activity and industrial enzymes. World J Microb Biotechnol. 2009;25:2103–11.
Nogi Y, Yoshizumi M, Miyazaki M. Thalassospira povalilytica sp. nov., a polyvinyl-alcohol-degrading marine bacterium. Int J Syst Evol Microbiol. 2014;64:1149–53.
Sedlacek P, Slaninova E, Enev V, Koller M, Nebesarova J, Marova I, et al. What keeps polyhydroxyalkanoates in bacterial cells amorphous? A derivation from stress exposure experiments. Appl Microbiol Biot. 2019;103:1905–17.
Steinbüchel A, Valentin HE. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett. 1995;128:219–28.
Madison LL, Huisman GW. Metabolic engineering of poly(3-hydroxyalkanoates): From DNA to plastic. Microbiol Mol Biol Rev 1999;63:21–53.
Kunioka M, Tamaki A, Doi Y. Crystalline and thermal properties of bacterial copolyesters: poly(3-hydroxybutyrate-co-3-hydroxyvalerate) and poly(3-hydroxybutyrate-co-4-hydroxybutyrate). Macromolecules. 1989;22:694–7.
Doi Y, Kitamura S, Abe H. Microbial synthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). Macromolecules. 1995;28:4822–8.
Abe H, Doi Y. Side-chain effect of second monomer units on crystalline morphology, thermal properties, and enzymatic degradability for random copolyesters of (R)-3-hydroxybutyric acid with (R)-3-hydroxyalkanoic acids. Biomacromolecules. 2002;3:133–8.
Shimamura E, Kasuya K, Kobayashi G, Shiotani T, Shima Y, Doi Y. Physical-properties and biodegradability of microbial poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). Macromolecules. 1994;27:878–80.
Suriyamongkol P, Weselake R, Narine S, Moloney M, Shah S. Biotechnological approaches for the production of polyhydroxyalkanoates in microorganisms and plants—a review. Biotechnol Adv. 2007;25:148–75.
Rehm BHA, Kruger N, Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis—the phaG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein coenzyme A transferase. J Biol Chem. 1998;273:24044–51.
Rehm BHA, Mitsky TA, Steinbüchel A. Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by pseudomonads: establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl Environ Microbiol. 2001;67:3102–9.
Fukui T, Shiomi N, Doi Y. Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J Bacteriol. 1998;180:667–73.
Chek MF, Kim SY, Mori T, Arsad H, Samian MR, Sudesh K, et al. Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics. Sci Rep. 2017;7:5312. https://doi.org/10.1038/s41598-017-05509-4.
Mezzolla V, D’Urso OF, Poltronieri P. Role of PhaC type I and type II enzymes during PHA biosynthesis. Polymer. 2018;10:910. https://doi.org/10.3390/polym10080910.
Rehm BHA. Polyester synthases: natural catalysts for plastics. Biochem J. 2003;376:15–33.
Tsuge T, Hyakutake M, Mizuno K. Class IV polyhydroxyalkanoate (PHA) synthases and PHA-producing Bacillus. Appl Microbiol Biotechnol. 2015;99:6231–40.
Lenz RW, Marchessault RH. Bacterial polyesters: biosynthesis, biodegradable plastics and biotechnology. Biomacromolecules. 2005;6:1–8.
Higuchi-Takeuchi M, Morisaki K, Numata K. A Screening method for the isolation of polyhydroxyalkanoate-producing purple non-sulfur photosynthetic bacteria from natural seawater. Front Microbiol. 2016;7:1509. https://doi.org/10.3389/fmicb.2016.01509.
Sabirova JS, Ferrer M, Regenhardt D, Timmis KN, Golyshin PN. Proteomic insights into metabolic adaptations in Alcanivorax borkumensis induced by alkane utilization. J Bacteriol. 2006;188:3763–73.
Shi XL, Wu YH, Jin XB, Wang CS, Xu XW. Alteromonas lipolytica sp. nov., a poly-beta-hydroxybutyrateproducing bacterium isolated from surface seawater. Int J Syst Evol Microbiol. 2017;67:237–42.
Mohandas SP, Balan L, Jayanath G, Anoop BS, Philip R, Cubelio SS, et al. Biosynthesis and characterization of polyhydroxyalkanoate from marine Bacillus cereus MCCB 281 utilizing glycerol as carbon source. Int J Biol Macromol. 2018;119:380–92.
Sathiyanarayanan G, Saibaba G, Kiran GS, Selvin J. Process optimization and production of polyhydroxybutyrate using palm jaggery as economical carbon source by marine sponge-associated Bacillus licheniformis MSBN12. Bioproc. Biosyst Eng. 2013;36:1817–27.
Lopez-Cortes A, Lanz-Landazuri A, Garcia-Maldonado JQ. Screening and isolation of PHB-producing bacteria in a polluted marine microbial mat. Microbiol Ecol. 2008;56:112–20.
Prabhu NN, Santimano MC, Mavinkurve S, Bhosle SN, Garg S. Native granule associated short chain length polyhydroxyalkanoate synthase from a marine derived Bacillus sp. NQ-11/A2. Anton Leeuw. 2010;97:41–50.
Hong JW, Song HS, Moon YM, Hong YG, Bhatia SK, Jung HR, et al. Polyhydroxybutyrate production in halophilic marine bacteria Vibrio proteolyticus isolated from the Korean peninsula. Bioprocess Biosyst Eng. 2019;42:603–10.
Kiran GS, Lipton AN, Priyadharshini S, Anitha K, Suarez LEC, Arasu MV, et al. Antiadhesive activity of poly-hydroxy butyrate biopolymer from a marine Brevibacterium casei MSI04 against shrimp pathogenic vibrios. Microb Cell Fact. 2014;13:114. https://doi.org/10.1186/s12934-014-0114-3.
Yamada M, Yukita A, Hanazumi Y, Yamahata Y, Moriya H, Miyazaki M, et al. Poly(3-hydroxybutyrate) production using mannitol as a sole carbon source by Burkholderia sp. AIU M5M02 isolated from a marine environment. Fish Sci. 2018;84:405–12.
Methe BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang XJ, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acid Sci USA. 2005;102:10913–8.
Numata K, Morisaki K, Tomizawa S, Ohtani M, Demura T, Miyazaki M. Synthesis of poly- and oligo(hydroxyalkanoate)s by deep-sea bacteria, Colwellia spp., Moritella spp., and Shewanella spp. Polym J. 2013;45:1094–100.
Hai T, Lange D, Rabus R, Steinbüchel A. Polyhydroxyalkanoate (PHA) accumulation in sulfate-reducing bacteria and identification of a class III PHA synthase (PhaEC) in Desulfococcus multivorans. Appl Environ Microbiol. 2004;70:4440–8.
Wang H, Tomasch J, Jarek M, Wagner-Dobler I. A dual-species co-cultivation system to study the interactions between Roseobacters and dinoflagellates. Front Microbiol. 2014;5:311. https://doi.org/10.3389/fmicb.2014.00311.
Xiao N, Jiao NZ. Formation of polyhydroxyalkanoate in aerobic anoxygenic phototrophic bacteria and its relationship to carbon source and light availability. Appl Environ Microbiol. 2011;77:7445–50.
Shrivastav A, Mishra SK, Shethia B, Pancha I, Jain D, Mishra S. Isolation of promising bacterial strains from soil and marine environment for polyhydroxyalkanoates (PHAs) production utilizing Jatropha biodiesel byproduct. Int J Biol Macromol. 2010;47:283–7.
Simon-Colin C, Raguénès G, Cozien J, Guezennec JG. Halomonas profundus sp. nov., a new PHA-producing bacterium isolated from a deep-sea hydrothermal vent shrimp. J Appl Microbiol. 2008;104:1425–32.
Lemechko P, Le Fellic M, Bruzaud S. Production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) using agro-industrial effluents with tunable proportion of 3-hydroxyvalerate monomer units. Int J Biol Macromol. 2019;128:429–34.
Han X, Satoh Y, Kuriki Y, Seino T, Fujita S, Suda T, et al. Polyhydroxyalkanoate production by a novel bacterium Massilia sp. UMI-21 isolated from seaweed, and molecular cloning of its polyhydroxyalkanoate synthase gene. J Biosci Bioeng. 2014;118:514–9.
Doronina NV, Trotsenko YA, Tourova TP. Methylarcula marina gen. nov., sp. nov. and Methylarcula terricola sp. nov.: novel aerobic, moderately halophilic, facultatively methylotrophic bacteria from coastal saline environments. Int J Syst Evol Microbiol. 2000;50:1849–59.
Liu XJ, Zhang J, Hong PH, Li ZJ. Microbial production and characterization of poly-3-hydroxybutyrate by Neptunomonas antarctica. Peerj. 2016;4:e2291. https://doi.org/10.7717/peerj.2291.
Cho JC, Giovannoni SJ. Oceanicola granulosus gen. nov., sp. nov. and Oceanicola batsensis sp. nov., poly-β-hydroxybutyrate-producing marine bacteria in the order ‘Rhodobacterales’. Int J Syst Evol Microbiol. 2004;54:1129–36.
Numata K, Morisaki K. Screening of marine bacteria to synthesize polyhydroxyalkanoate from lignin: contribution of lignin derivatives to biosynthesis by Oceanimonas doudoroffii. ACS Sustain Chem Eng. 2015;3:569–73.
Boyandin AN, Kalacheva GS, Rodicheva EK, Volova TG. Synthesis of reserve polyhydroxyalkanoates by luminescent bacteria. Microbiology. 2008;77:318–23.
Wang Q, Zhang HX, Chen Q, Chen XL, Zhang YZ, Qi QS. A marine bacterium accumulates polyhydroxyalkanoate consisting of mainly 3-hydroxydodecanoate and 3-hydroxydecanoate. World J Microbiol Biotechnol. 2010;26:1149–53.
Simon-Colin C, Alain K, Colin S, Cozien J, Costa B, Guezennec JG, et al. A novel mcl PHA-producing bacterium, Pseudomonas guezennei sp. nov., isolated from a ‘kopara’ mat located in Rangiroa, an atoll of French Polynesia. J Appl Microbiol. 2008;104:581–6.
Jamil N, Ahmed N, Edwards DH. Characterization of biopolymer produced by Pseudomonas sp. CMG607w of marine origin. J Gen Appl Microbiol. 2007;53:105–9.
Higuchi-Takeuchi M, Numata K. Acetate-inducing metabolic states enhance polyhydroxyalkanoate production in marine purple non-sulfur bacteria under aerobic conditions. Front Bioeng Biotechnol. 2019;7:118. https://doi.org/10.3389/fbioe.2019.00118.
Higuchi-Takeuchi M, Morisaki K, Toyooka K, Numata K. Synthesis of high-molecular-weight polyhydroxyalkanoates by marine photosynthetic purple bacteria. PLoS ONE. 2016;11. https://doi.org/10.1371/journal.pone.0160981.
Gonzalez-Garcia Y, Nungaray J, Cordova J, Gonzalez-Reynoso O, Koller M, Atlic A, et al. Biosynthesis and characterization of polyhydroxyalkanoates in the polysaccharide-degrading marine bacterium Saccharophagus degradans ATCC 43961. J Ind Microbiol Biotechnol. 2008;35:629–33.
Ting L, Williams TJ, Cowley MJ, Lauro FM, Guilhaus M, Raftery MJ, et al. Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ Microbiol. 2010;12:2658–76.
Shrivastav A, Mishra SK, Mishra S. Polyhydroxyalkanoate (PHA) synthesis by Spirulina subsalsa from Gujarat coast of India. Int J Biol Macromol. 2010;46:255–60.
Sasidharan RS, Bhat SG, Chandrasekaran M. Biocompatible polyhydroxybutyrate (PHB) production by marine Vibrio azureus BTKB33 under submerged fermentation. Ann Microbiol. 2015;65:455–65.
Mohandas SP, Balan L, Lekshmi N, Cubelio SS, Philip R, Bright SIS. Production and characterization of polyhydroxybutyrate from Vibrio harveyi MCCB 284 utilizing glycerol as carbon source. J Appl Microbiol. 2017;122:698–707.
Wei YH, Chen WC, Wu HS, Janarthanan OM. Biodegradable and biocompatible biomaterial, polyhydroxybutyrate, produced by an indigenous Vibrio sp. BM-1 isolated from marine environment. Mar Drugs. 2011;9:615–24.
Numata K, Doi Y. Biosynthesis of Polyhydroxyalkanaotes by a novel facultatively anaerobic Vibrio sp. under marine conditions. Mar Biotechnol. 2012;14:323–31.
Stabili L, Cardone F, Alifano P, Tredici SM, Piraino S, Corriero G, et al. Epidemic Mortality of the sponge Ircinia variabilis (Schmidt, 1862) associated to proliferation of a Vibrio Bacterium. Microb Ecol. 2012;64:802–13.
Foong CP, Lau NS, Deguchi S, Toyofuku T, Taylor TD, Sudesh K, et al. Whole genome amplification approach reveals novel polyhydroxyalkanoate synthases (PhaCs) from Japan Trench and Nankai Trough seawater. BMC Microbiol. 2014;14:318. https://doi.org/10.1186/s12866-014-0318-z.
Doi Y, Kanesawa Y, Tanahashi N, Kumagai Y. Biodegradation of microbial polyesters in the marine-environment. Polym Degrad Stabil. 1992;36:173–7.
Tsuji H, Suzuyoshi K. Environmental degradation of biodegradable polyesters 2. Poly(ε-caprolactone), poly[(R)-3-hydroxybutyrate], and poly(L-lactide) films in natural dynamic seawater. Polym Degrad Stabil. 2002;75:357–65.
Tsuji H, Suzuyoshi K. Environmental degradation of biodegradable polyesters 1. Poly(ε-caprolactone), poly[(R)-3-hydroxybutyrate], and poly(L-lactide) films in controlled static seawater. Polym Degrad Stabil. 2002;75:347–55.
Rutkowska M, Krasowska K, Heimowska A, Adamus G, Sobota M, Musiol M, et al. Environmental degradation of blends of atactic poly (R,S)-3-hydroxybutyrate with natural PHBV in Baltic sea water and compost with activated sludge. J Polym Environ. 2008;16:183–91.
Volova TG, Boyandin AN, Vasil’ev AD, Karpov VA, Kozhevnikov IV, Prudnikova SV, et al. Biodegradation of polyhydroxyalkanoates (PHAs) in the South China Sea and identification of PHA-degrading bacteria. Microbiology. 2011;80:252. https://doi.org/10.1134/S0026261711020184.
Thellen C, Coyne M, Froio D, Auerbach M, Wirsen C, Ratto JA. A processing, characterization and marine biodegradation study of melt-extruded polyhydroxyalkanoate (PHA) films. J Polym Environ. 2008;16:1–11.
Deroine M, Cesar G, Le Duigou A, Davies P, Bruzaud S. Natural degradation and biodegradation of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in liquid and solid marine environments. J Polym Environ. 2015;23:493–505.
Imam SH, Gordon SH, Shogren RL, Tosteson TR, Govind NS, Greene RV. Degradation of starch-poly(beta-hydroxybutyrate-co-beta-hydroxyvalerate) bioplastic in tropical coastal waters. Appl Environ Microbiol. 1999;65:431–7.
Suzuki M, Tachibana Y, Kazahaya J, Takizawa R, Muroi F, Kasuya K. Difference in environmental degradability between poly(ethylene succinate) and poly(3-hydroxybutyrate). J Polym Res. 2017;24:217. https://doi.org/10.1007/s10965-017-1383-4.
Dilkes-Hoffman LS, Lant PA, Laycock B, Pratt S. The rate of biodegradation of PHA bioplastics in the marine environment: a meta-study. Mar Pollut Bull. 2019;142:15–24.
Zettler ER, Mincer TJ, Amaral-Zettler LA. Life in the “Plastisphere”: microbial communities on plastic marine debris. Environ Sci Technol. 2013;47:7137–46.
Morohoshi T, Ogata K, Okura T, Sato S. Molecular characterization of the bacterial community in biofilms for degradation of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) films in seawater. Microbes Environ. 2018;33:19–25.
Pinnell LJ, Turner JW. Shotgun metagenomics reveals the benthic microbial community response to plastic and bioplastic in a coastal marine environment. Front Microbiol. 2019;10:1252. https://doi.org/10.3389/fmicb.2019.01252.
Zadjelovic V, Chhun A, Quareshy M, Silvano E, Hernandez-Fernaud JR, Aguilo-Ferretjans MM, et al. Beyond oil degradation: enzymatic potential of Alcanivorax to degrade natural and synthetic polyesters. Environ Microbiol. 2020;22:1356–69.
Kita K, Ishimaru K, Teraoka M, Yanase H, Kato N. Properties of poly(3-hydroxybutyrate) depolymerase from a marine bacterium, Alcaligenes faecalis AE122. Appl Environ Microbiol. 1995;61:1727–30.
Mergaert J, Wouters A, Anderson C, Swings J. In-situ biodegradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in natural-waters. Can J Microbiol. 1995;41:154–9.
Kato C, Honma A, Sato S, Okura T, Fukuda R, Nogi Y. Poly 3-hydroxybutyrate-co-3-hydroxyhexanoate films can be degraded by the deep-sea microbes at high pressure and low temperature conditions. High Press Res. 2019;39:248–57.
Volova TG, Boyandin AN, Vasiliev AD, Karpov VA, Prudnikova SV, Mishukova OV, et al. Biodegradation of polyhydroxyalkanoates (PHAs) in tropical coastal waters and identification of PHA-degrading bacteria. Polym Degrad Stabil. 2010;95:2350–9.
Ma WT, Lin JH, Chen HJ, Chen SY, Shaw GC. Identification and characterization of a novel class of extracellular poly(3-hydroxybutyrate) depolymerase from Bacillus sp Strain NRRL B-14911. Appl Environ Microbiol. 2011;77:7924–32.
Kasuya K, Mitomo H, Nakahara M, Akiba A, Kudo T, Doi Y. Identification of a marine benthic P(3HB)-degrading bacterium isolate and characterization of its P(3HB) depolymerase. Biomacromolecules. 2000;1:194–201.
Ghanem NB, Mabrouk MES, Sabry SA, El-Badan DES. Degradation of polyesters by a novel marine Nocardiopsis aegyptia sp. nov.: Application of Plackett-Burman experimental design for the improvement of PHB depolymerase activity. J Gen Appl Microbiol. 2005;51:151–8.
Leathers TD, Govind NS, Greene RV. Biodegradation of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by a tropical marine bacterium, Pseudoalteromonas sp. NRRL b-30083. J Polym Environ. 2000;8:119–24.
Uefuji M, Kasuya K, Doi Y. Enzymatic degradation of poly (R)-3-hydroxybutyrate: secretion and properties of PHB depolymerase from Pseudomonas stutzeri. Polym Degrad Stabil. 1997;58:275–81.
Sung CC, Tachibana Y, Suzuki M, Hsieh WC, Kasuya K. Identification of a poly(3-hydroxybutyrate)-degrading bacterium isolated from coastal seawater in Japan as Shewanella sp. Polym Degrad Stabil. 2016;129:268–74.
Mabrouk MM, Sabry SA. Degradation of poly (3-hydroxybutyrate) and its copolymer poly (3-hydroxybutyrate-co-3-hydroxyvalerate) by a marine Streptomyces sp. SNG9. Microbiol Res. 2001;156:323–35.
Kita K, Mashiba S, Nagita M, Ishimaru K, Okamoto K, Yanase H, et al. Cloning of poly(3-hydroxybutyrate) depolymerase from a marine bacterium, Alcaligenes faecalis AE122, and characterization of its gene product. BBA-Gene Struct Expr. 1997;1352:113–22.
Kasuya K, Takano T, Tezuka Y, Hsieh WC, Mitomo H, Doi Y. Cloning, expression and characterization of a poly(3-hydroxybutyrate) depolymerase from Marinobacter sp. NK-1. Int J Biol Macromol. 2003;33:221–6.
Ohura T, Kasuya K, Doi Y. Cloning and characterization of the polyhydroxybutyrate depolymerase gene of Pseudomonas stutzeri and analysis of the function of substrate-binding domains. Appl Environ Microbiol. 1999;65:189–97.
Hisano T, Kasuya K, Tezuka Y, Ishii N, Kobayashi T, Shiraki M, et al. The crystal structure of polyhydroxybutyrate depolymerase from Penicillium funiculosum provides insights into the recognition and degradation of biopolyesters. J Mol Biol. 2006;356:993–1004.
Kasuya K, Inoue Y, Tanaka T, Akehata T, Iwata T, Fukui T, et al. Biochemical and molecular characterization of the polyhydroxybutyrate depolymerase of Comamonas acidovorans YM1609, isolated from freshwater. Appl Environ Microbiol. 1997;63:4844–52.
Sung CC, Tachibana Y, Kasuya K. Characterization of a thermolabile poly(3-hydroxybutyrate) depolymerase from the marine bacterium Shewanella sp. JKCM-AJ-6,1α. Polym Degrad Stabil. 2016;129:212–21.
NOAA. Sea surface temperature (sst) contour charts. 2020. https://www.ospo.noaa.gov/Products/ocean/sst/contour/.
Kroeker KJ, Kordas RL, Crim R, Hendriks IE, Ramajo L, Singh GS, et al. Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob Change Biol. 2013;19:1884–96.
Xue DW, Zhang XQ, Lu XL, Chen G, Chen ZH. Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front Plant Sci. 2017;8:621. https://doi.org/10.3389/fpls.2017.00621.
Yeats TH, Rose JKC. The formation and function of plant cuticles. Plant Physiol. 2013;163:5–20.
Philippe G, Sørensen I, Jiao C, Sun X, Fei Z, Domozych DS, et al. Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr Opin Plant Biol. 2020;55:11–20.
Broder L, Tesi T, Salvado JA, Semiletov IP, Dudarev OV, Gustafsson O. Fate of terrigenous organic matter across the Laptev Sea from the mouth of the Lena River to the deep sea of the Arctic interior. Biogeosciences. 2016;13:5003–19.
Broder L, Tesi T, Andersson A, Eglinton TI, Semiletov IP, Dudarev OV, et al. Historical records of organic matter supply and degradation status in the East Siberian Sea. Org Geochem. 2016;91:16–30.
Goni MA, Yunker MB, Macdonald RW, Eglinton TI. Distribution and sources of organic biomarkers in arctic sediments from the Mackenzie River and Beaufort Shelf. Mar Chem. 2000;71:23–51.
Prahl FG, Ertel JR, Goni MA, Sparrow MA, Eversmeyer B. Terrestrial organic-carbon contributions to sediments on the Washington margin. Geochim Cosmochim Acta. 1994;58:3035–48.
Kaal J, Serrano O, Cortizas AM, Baldock JA, Lavery PS. Millennial-scale changes in the molecular composition of Posidonia australis seagrass deposits: Implications for Blue Carbon sequestration. Org Geochem. 2019;137:103898. https://doi.org/10.1016/j.orggeochem.2019.07.007.
Moran KL, Bjorndal KA. Simulated green turtle grazing affects nutrient composition of the seagrass Thalassia testudinum. Mar Biol. 2007;150:1083–92.
Olsen JL, Rouze P, Verhelst B, Lin YC, Bayer T, Collen J, et al. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature. 2016;530:331–5.
Sugiura H, Kawasaki Y, Suzuki T, Maegawa M. The structural and histochemical analyses and chemical characters of the cuticle and epidermal walls of cotyledon in ungerminated seeds of Zostera marina L. Fish Sci. 2009;75:369–77.
Lu B, Wang GX, Huang D, Ren ZL, Wang XW, Zhen ZC, et al. Comparison of PCL degradation in different aquatic environments: effects of bacteria and inorganic salts. Polym Degrad Stabil. 2018;150:133–9.
Molitor R, Bollinger A, Kubicki S, Loeschcke A, Jaeger KE, Thies S. Agar plate-based screening methods for the identification of polyester hydrolysis by Pseudomonas species. Microb Biotechnol. 2020;13:274–84.
Suzuki M, Tachibana Y, Oba K, Takizawa R, Kasuya K. Microbial degradation of poly(ε-caprolactone) in a coastal environment. Polym Degrad Stabil. 2018;149:1–8.
Almeida EL, AFC Rincon, Jackson SA, ADW Dobson. In silico screening and heterologous expression of a polyethylene terephthalate hydrolase (PETase)-like enzyme (SM14est) with polycaprolactone (PCL)-degrading activity, from the marine sponge-derived strain Streptomyces sp. SM14. Front Microbiol. 2019;10:2187. https://doi.org/10.3389/fmicb.2019.02187.
Sekiguchi T, Sato T, Enoki M, Kanehiro H, Uematsu K, Kato C. Isolation and characterization of biodegradable plastic degrading bacteria from deep-sea environments. JAMSTEC Rep. Res Dev. 2011;11:33–41.
Kovacic F, Babic N, Krauss U, Jaeger K. Classification of lipolytic enzymes from bacteria. In: Handbook of hydrocarbon and lipid microbiology. Cham: Springer; 2019;24. https://doi.org/10.1007/978-3-319-39782-5_39-1.
Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343:177–83.
Nikolaivits E, Kanelli M, Dimarogona M, Topakas E. A middle-aged enzyme still in its prime: recent advances in the field of cutinases. Catalysts. 2018;8:612. https://doi.org/10.3390/catal8120612.
Chen S, Su LQ, Chen J, Wu J. Cutinase: characteristics, preparation, and application. Biotechnol Adv. 2013;31:1754–67.
Chen S, Tong X, Woodard RW, Du GC, Wu J, Chen J. Identification and characterization of bacterial cutinase. J Biol Chem. 2008;283:25854–62.
Reis P, Holmberg K, Watzke H, Leser ME, Miller R. Lipases at interfaces: a review. Adv Colloid Interfac. 2009;147:237–50.
Zumstein MT, Rechsteiner D, Roduner N, Perz V, Ribitsch D, Guebitz GM, et al. Enzymatic hydrolysis of polyester thin films at the nanoscale: effects of polyester structure and enzyme active-site accessibility. Environ Sci Technol. 2017;51:7476–85.
Shi K, Jing J, Song L, Su TT, Wang ZY. Enzymatic hydrolysis of polyester: Degradation of poly(ε-caprolactone) by Candida antarctica lipase and Fusarium solani cutinase. Int J Biol Macromol. 2020;144:183–9.
Trodler P, Pleiss J. Modeling structure and flexibility of Candida antarctica lipase B in organic solvents. BMC Struct Biol. 2008;8:9. https://doi.org/10.1186/1472-6807-8-9.
Hajighasemi M, Nocek BP, Tchigvintsev A, Brown G, Flick R, Xu XH, et al. Biochemical and structural insights into enzymatic depolymerization of polylactic acid and other polyesters by microbial carboxylesterases. Biomacromolecules. 2016;17:2027–39.
Danso D, Schmeisser C, Chow J, Zimmermann W, Wei R, Leggewie C, et al. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl Environ Microbiol. 2018;84:e02773–17. https://doi.org/10.1128/AEM.02773-17.
Bollinger A, Thies S, Knieps-Grunhagen E, Gertzen C, Kobus S, Hoppner A, et al. A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri - Structural and functional insights. Front Microbiol. 2020;11:114. https://doi.org/110.3389/fmicb.2020.00114.
Shinomiya M, Iwata T, Kasuya K, Doi Y. Cloning of the gene for poly(3-hydroxybutyric acid) depolymerase of Comamonas testosteroni and functional analysis of its substrate-binding domain. FEMS Microbiol Lett. 1997;154:89–94.
Acknowledgements
KK is grateful for financial supports from JST-Mirai Program grant number JP19217252 and JSPS KAKENHI grant number JP19H04311.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Suzuki, M., Tachibana, Y. & Kasuya, Ki. Biodegradability of poly(3-hydroxyalkanoate) and poly(ε-caprolactone) via biological carbon cycles in marine environments. Polym J 53, 47–66 (2021). https://doi.org/10.1038/s41428-020-00396-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41428-020-00396-5
This article is cited by
-
Hydrolyzable and biocompatible aliphatic polycarbonates with ether-functionalized side chains attached via amide linkers
Polymer Journal (2024)
-
Characteristics of the steam degradation of poly(lactic acid) and poly(3-hydroxybutyrate-co-3-hydroxyhexanoate)
Polymer Journal (2024)
-
Microbial decomposition of biodegradable plastics on the deep-sea floor
Nature Communications (2024)
-
Promoted biodegradation behavior of poly(L-lactic acid) in seawater conditions through blending amorphous polyhydroxyalkanoate
Macromolecular Research (2024)
-
Biodegradable microplastics interaction with pollutants and their potential toxicity for aquatic biota: a review
Environmental Chemistry Letters (2024)