Abstract
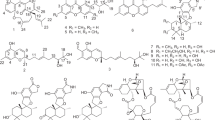
Five new prenylated indole alkaloids, 17-hydroxynotoamide D (1), 17-O-ethylnotoamide M (2), 10-O-acetylsclerotiamide (3), 10-O-ethylsclerotiamide (4), and 10-O-ethylnotoamide R (5) were isolated from a co-culture of marine-derived fungi Aspergillus sulphureus KMM 4640 and Isaria felina KMM 4639. The structures of 1−5 were determined by detailed analysis of spectroscopic data and by comparison with related known compounds. The absolute configurations of 1−5 were determined by time-dependent density functional theory (TD-DFT) calculations of ECD spectra. Compound 2 is able to inhibit the colony formation of human prostate cancer cells 22Rv1 at non-cytotoxic concentration of 10 μM.
Similar content being viewed by others
Introduction
Fungi isolated from marine environments have received great attention as prolific sources of new bioactive secondary metabolites [1,2,3]. Recent studies show that one of the factors causing the induction of biosynthesis of new metabolites from marine fungi is co-culturing with other microorganisms, including fungi. For example, in fungus-bacterium fermentation, cytotoxic phenolic polyketides were isolated from co-cultivation of marine-derived Penicillium sp. and Streptomyces fradiae [4]. Antimicrobial citrifelins A and B with a unique tetracyclic framework were characterized from a co-culture of two marine-derived fungi Penicillium citrinum and Beauveria felina [5] and antifungal cyclic tetrapeptide was produced by co-cultivation of two marine mangrove fungi Phomopsis sp. and Alternaria sp. [6].
In previously published works, we described isolation and identification of ten new highly oxygenated chromene derivatives—oxirapentyns B–K, pyran polyketide–isariketide A, benzofuran–acremine S from the marine sediment-derived fungus Isaria felina KMM 4639 [7,8,9], and new decaline derivative—decumbenone C together with known compounds decumbenones A and B, diorcinol, and brevianamide F from fungus Aspergillus sulphureus KMM 4660 [10]. After that, we investigated the secondary metabolites produced by their co-cultivation, which yielded new diorcinol J and oxirapentyn L [11, 12]. Herein, we report the isolation, structure elucidation, and biological assay results of five new prenylated indole alkaloids notoamide family 1‒5 (Fig. 1) together with known compounds (‒)-notoamide B (6), notoamide C (7), dehydronotoamide C (8), notoamide D (9), notoamide F (10), notoamide Q (11), 17-epi-notoamide Q (12), notoamide M (13), and sclerotiamide (14), which were produced in response to the addition of a marine fungus Isaria felina to an established 7-day-old culture of a marine-derived fungus Aspergillus sulphureus.
Results and discussion
The molecular formula of 1 was determined to be C26H31N3O5 from a HRESIMS peak at m/z 466.2341 [M + H]+and was in accordance with 13C NMR data. A close inspection of 1H and 13C NMR data of 1 by DEPT and HSQC (Tables 1 and 2, Supporting Information, Figure S4) indicated the presence of five olefinic methines (δH 5.54, 6.28, 6.29, 6.30, 7.00, δC 129.3, 108.6, 116.7, 146.6, 123.7), one olefinic methylene (δH 5.04 and 5.07, δC 112.9), four methyl groups (δH 1.25, 1.26, 1.38, 1.40, δC 26.4, 23.4, 28.0, 27.5), two amide carbonyls (δC 167.4 and 171.2), one sp2- (δC 154.8) and three sp3 (δC 76.3, 88.9 and 90.3) oxygen-bearing carbons, three sp2 (δC 106.0, 122.7, 145.4), and two sp3 (δC 45.3, 91.7) quaternary carbons along with one methine and four methylene groups.
The 13C NMR data (Table 2) for this compound were very similar to those obtained for notoamide D [13] with the exception of C-11 and C-15–C-18 carbon signals of the diketopiperazine moiety. The molecular mass difference of 16 mass units between 1 and notoamide D and the HMBC correlations from H-15a,b to C-16 (δC 37.9), C-17 (δC 90.3) and from H-16b to C-14 (δC 44.8), C-15 (δC 20.7), C-16, C-17, and C-18 (δC 171.2) established the location of the alcohol function at C-17. The molecular structure and relative configuration of 1 were unequivocally confirmed by X-ray analysis, which was carried out for a single crystal the composition (C26H31N3O5))2·3H2O, obtained by recrystallization from acetone-water (Supporting Information, Figure S7) (Fig. 2).
The absolute configuration of 1 was determined by a combination of X-ray analysis and ECD spectroscopy data. The geometry of 1, obtained in the X-ray experiment, was taken as a starting point for the following conformational analysis, performed using density functional theory (DFT) and polarization continuum model (PCM) (Supporting Information, Figure S40). The internal rotations of the substituents at C-2, C-3, and C-17, as well as the rotation of the vinyl group were accounted for. It was found that nearly 80% of compound 1 molecules when solvated in CH3OH solvent exist in two rotameric forms (S1-A and S1-B, see Supporting Information, Figure S33), in which the intramolecular bond of the OH…π type is formed [14, 15]. The “cis” orientation of the vinyl and methyl groups relative to C-21−C-22 bond is preferable.
The conformational analysis is a base for the further calculations of different spectroscopic properties. Based on the conformational analysis results, a number of most stable conformations may be selected, for which the UV, ECD, and other spectra must be calculated and statistically averaged. In present work we chosen the condition ΔGim ≤ 3 kcal mol−1 (for details see Supporting Information, Figure S40). The excitation energies and the rotatory strengths were calculated using time-dependent density functional theory (TD-DFT) with B3LYP exchange-correlation functional and 6–31 G(d) basis set. One can see from Figure S32 (Supporting Information) that this level of quantum-chemical modeling is tolerable to reproduce correctly the positions of the absorbance bands in the experimental UV spectrum for 1.
The comparison of statistically averaged theoretical and experimental ECD spectra for 1 is presented on Fig. 3. Both spectra are qualitatively similar in the representative region above 200 nm, which proves the 2S,3R,11S,17R absolute configuration for 1. Compound 1 was named 17-hydroxynotoamide D.
The molecular formula of 2 was determined to be C28H35N3O5 from a HRESIMS peak at m/z 516.2469 [M + Na]+ and was in accordance with 13C NMR data. The general features of the 1H and 13C NMR spectra (Tables 1 and 2) of 2 matched those reported for notoamide M [16] and notoamide Q [17]. The correlations observed in the COSY spectrum and HMBC correlations from H3-31 to C-30 (δC 58.8) and from H2-30 to C-17 (δC 90.4) revealed the location of an ethyl ether at C-17 in 2.
The NOE correlations H-11/H2-30, H3-31 in 2 indicated that the diketopiperazine ring is of cis configuration. Compound 2 exhibited a nearly identical ECD spectrum to those of notoamides C, M, and Q [18] (Supporting Information, Figure S34), establishing its absolute configuration as 3S,11S,17R. Compound 2 was named 17-O-ethylnotoamide M.
The molecular formula of 3 was determined to be C28H31N3O6 from a HRESIMS peak at m/z 528.2112 [M + Na]+ and was in accordance with 13C NMR data (Table 2). The 1H and 13C NMR data for 3 were in a good agreement with those for sclerotiamide [19] with the exception of H-10 signal. The HMBC correlations (Supporting Information, Fig. S18) together with the molecular mass difference of 42 mass units between 3 and sclerotiamide indicated the presence of an acetoxy group at C-10 in 3.
The NOE correlations H-21/NH-19, H3-23, and H-10/H-4, H3-24 determined the relative configuration of 3. The ECD spectrum of 3 showed characteristic Cotton effects at λ202–11.12, λ225 + 7.96, and λ240–3.56 (Supporting Information, Figure S35), which were in good agreement with those for notoamide H [20]. Thus, the absolute configuration of 3 was established as 3R,10S,11R,17S,21S. Compound 3 was named 10-O-acetylsclerotiamide.
The molecular formula of compound 4 was determined as C28H33N3O5 from a HRESIMS peak at m/z 514.2316 [M + Na]+ and was in accordance with 13C NMR data. The 13C NMR data for this compound were very similar to those obtained for 3 with the exception of C-10 carbon signal. The correlations observed in the COSY spectrum and HMBC correlations from H3-31 to C-30 (δC 67.9) and from H2-30 to C-10 (δC 80.3) revealed the location of an ethyl ether at C-10 in 4.
The relative configuration of 4 was defined based on observed NOE correlations between H-21 and H3-23, NH-19 and between H-10 and H3-24, H-4. Compound 4 exhibited a nearly identical CD spectrum to that of 3 (Supporting Information, Figure S35), establishing its absolute configuration as 3R,10S,11R,17S,21S. Compound 4 was named 10-O-ethylsclerotiamide.
The molecular formula of compound 5 was determined as C28H33N3O4 from a HRESIMS peak at m/z 498.2332 [M + Na]+ and was in accordance with 13C NMR data. The 1H and 13C NMR data (Tables 1 and 2) observed for the 5 closely resembled those of notoamide F [20] with the exception of C-10 carbon signal. The structure and location of the ethyl ester at C-10 were established as for 4.
The ECD spectrum of 5 showed the characteristic Cotton effects at λ203–16.25, λ224 + 16.59, and λ247–1.77 (Supporting Information, Figure S36). These CEs were in good agreement with those for notoamide F [20], which allowed us to determine the absolute configuration of 5 as 10S,11R,17S,21S. Compound 5 was named 10-O-ethylnotoamide R.
Furthermore, we observed compounds 2 and 4 in original EtOAc extract of co-culture A. sulphureus and I. felina by HPLC-MS method (Supporting Information, Figure S37). 10-O-ethyl ester notoamide R (5) has not been detected from original extract, thus 5 might be not a natural product and obtained during isolation process.
Besides the new compounds, (‒)-notoamide B (6) [13], notoamide C (7) [13], dehydronotoamide C (8) [18], notoamide D (9) [13], notoamide F (10) [20], notoamide Q (11) [17], 17-epi-notoamide Q (12) [18], notoamide M (13) [16], and sclerotiamide (14) [19] (Supporting Information, Figure S38) were also isolated from the co-culture extract.
We have investigated the effect of the substances 1, 2, 5‒9, 13, and 14 on the viability of human non-malignant and prostate cancer cells, as well as on colony formation of human prostate cancer cells 22Rv1. MTT assay revealed all the substances tested to be non-cytotoxic to non-malignant (MRC-9 and HEK 293 T), as well as malignant (22Rv1, PC-3, and LNCaP) cell lines at concentrations up to 100 µM after 48 h of treatment (data not shown). At the same time, compounds 2, 6, 8, 13, and 14 inhibited the colony formation of 22Rv1 prostate cancer cells at a concentration of 100 µM (Fig. 4). Of note, compounds 2 and 13 significantly decreased colony formation at concentrations of 10 µM by 25 and 55%, respectively. 22Rv1 cells are known to be resistant to hormone therapy including the novel 2nd generation drugs abiraterone and enzalutamide due to the presence of androgen receptor splice variant AR-V7 [21]. Therefore drugs, which are active in these cells, may be may be of interest to be further investigated in the therapy of human drug-resistant prostate cancer. Thus, further evaluation of these compounds in different human prostate cancer cell lines is planned.
Effect of compounds on colony formation of 22Rv1 cells. Cells were treated with the drugs for 48 h. 100 viable cells were plated in the wells of 6-well plates and incubated for 10 days. Cell colonies were fixed, stained, and counted with the naked eye. Significant difference from the control are shown as follows: *p < 0.05 (Student’s t-test)
In conclusion, it should be noted that the investigated fungi, cultured alone, did not produce these metabolites. However, brevianamide F, which is a common precursor of the obtained alkaloids, was isolated earlier from the fungus A. sulphureus [22].
Materials and methods
General experimental procedures
Optical rotations were measured on a Perkin-Elmer 343 polarimeter in CHCl3. UV spectra were recorded on a Shimadzu UV-1601PC spectrometer in MeOH. ECD spectra were measured with a Chirascan-Plus CD spectrometer (Leatherhead, UK) in MeOH. 1H and 13C NMR spectra were recorded in CDCl3 on a Bruker Avance-500 and Avance III-700 spectrometers operating at 500.13 MHz and 125.77 MHz, and 700.13 and 176.04 MHz, respectively, using TMS as an internal standard. HRESIMS spectra were obtained on a Maxis Impact II Q-TOF (Bruker, Germany) mass spectrometer. Low pressure liquid column chromatography was performed using Si gel L (40/100 μm, Sorbpolimer, Russia) and Sephadex LH-20 (GE Healthcare, Sweden). Plates (4.5 × 6.0 cm) precoated with Si gel (5-17 µm, Sorbfil) were used for TLC. Preparative HPLC was carried out on a Beckman–Altex chromatograph, using a YMC ODS-AM (5 µm, 10 × 250 mm) column with an RIDK–122 refractometer and Agilent 1100 Series chromatograph, using Beckman Ultrashere Si (5 μm, 4.0 × 250 mm) and Supelco Discovery C-18 (5 μm, 4.6 × 250 mm) columns with an RID–G1362A refractometer.
Fungal strain
The isolation and identification of the both fungi were described previously [7, 10]. Strains are stored at Collection of Marine Microorganisms, PIBOC, Vladivostok, Russia, with the codes KMM 4640 for Aspergillus sulpureus and KMM 4639 for Isaria felina.
Fermentation, extraction, and isolation
The fungi A. sulphureus and I. felina were cultivated separately at 22 °C for 7 days in Erlenmeyer flasks (500 mL) each containing 20 g of rice, 20 mg of yeast extract, 10 mg of KH2PO4, and 40 mL of natural sea water. After that, I. felina mycelium was inoculated into 20 flasks with A. sulphureus culture. Then fungal cultures were co-cultivated for 14 days. At the end of the incubation period, the mycelium and medium were homogenized and extracted with EtOAc (2 L). The extract was concentrated to dryness. The residue was dissolved in 20% MeOH–H2O (1 L) and was extracted with n-hexane (0.2 L × 3) and EtOAc (0.2 L × 3). After evaporation of the EtOAc layer, the residual material (3.7 g) was passed over silica columns (4 × 20 cm), which was eluted first with n-hexane (1 L) followed by a step gradient from 5 to 100% EtOAc in n-hexane (total volume 7 L). Fractions of 200 mL were collected and combined on the basis on TLC (Si gel, toluene–isopropanol 6:1, v/v).
The n-hexane–EtOAc (85:15) eluate (235 mg) was purified by RP HPLC on a YMC ODS-AM column using MeOH–H2O (85:15) to yield individual compound 1 (62 mg), 9 (20 mg), and fraction A (50 mg). Fraction A was purified by NP HPLC on an Ultrasphere Si column eluted with CHCl3–EtOH (100:5) to yield 8 (5 mg) and 11 (2 mg).
The n-hexane–EtOAc (75:25) eluate (550 mg) was purified by RP HPLC on a YMC ODS-AM column using MeOH–H2O (80:20) to yield fractions B (17 mg), C (20 mg), D (5 mg), and E (25 mg). Fractions B was purified by NP HPLC on an Ultrasphere Si column eluted with EtOAc–CHCl3–EtOH (100:100:4) to yield 2 (4.5 mg) and EtOAc–CHCl3–EtOH (100:50:3) to yield 13 (2.1 mg). Fractions C was purified by NP HPLC on an Ultrasphere Si column eluted with CHCl3–n-hexane–EtOH (100:25:3) to yield 3 (0.5 mg), 5 (4.5 mg), and 14 (2.3 mg). Fractions D was purified by NP HPLC on an Ultrasphere Si column eluted with CHCl3–n-hexane–EtOH (100:20:3) to yield 4 (0.5 mg) and 10 (1 mg). Fractions E was purified by RP HPLC on a Supelco Discovery C-18 column eluting with CH3CN–H2O (45:55) to yield 6 (0.5 mg).
The n-hexane–EtOAc (50:50) eluate (140 mg) was purified by RP HPLC on a YMC ODS-AM column using MeOH–H2O (80:20) to yield fraction F (12 mg). Fraction F was purified by RP HPLC on a Supelco Discovery C-18 column eluting with CH3CN–H2O (45:55) to yield 7 (2 mg) and 12 (0.2 mg).
For HPLC-MS analyses, the original EtOAc extract was purified by RP HPLC on a Supelco Discovery C-18 column using MeOH–H2O (85:15).
17-hydroxynotoamide D (1): colorless crystal; mp 168‒170 °C; [α]20D –115 (c 0.07, MeOH); UV (MeOH) λmax (log ε) 329 (3.66), 286 (3.89), 236 (4.38), 196 (4.59) nm; CD (0.22 mM, MeOH) λmax (Δε) 221 (–28.53), 239 (+0.86), 255 (–21.00), 333 (+1.90) nm; IR (CDCl3) νmax 2977, 1676, 1645, 1458, 1382 cm−1; 1H and 13C NMR data, see Tables 1 and 2, Supporting Information S1–S6; HRESIMS m/z 466.2341 [M + H]+ (calcd for C26H32N3O5, 466.2336); 488.2166 [M + Na]+ (calcd for C26H31N3O5Na, 488.2156).
17-O-ethylnotoamide M (2): white powder, [α]20D + 79 (c 0.08, MeOH); UV (MeOH) λmax (log ε) 315 (3.31), 247 (4.43), 196 (4.56) nm; CD (0.24 mM, MeOH) λmax (Δε) 200 (–22.16), 231 (+15.14), 252 (–6.63), 276 (+7.14), 320 (+1.13) nm; IR (CDCl3) νmax 2974, 1702, 1679, 1645, 1459, 1381 cm−1; 1H and 13C NMR data, see Tables 1 and 2, Supporting Information S8–S13; HRESIMS m/z 516.2469 [M + Na]+ (calcd for C28H35N3O5Na, 516.2468).
10-O-acetylsclerotiamide (3): white powder, [α]20D –35 (c 0.04, MeOH); UV (MeOH) λmax (log ε) 243 (4.32), 197 (4.33) nm; CD (0.49 mM, MeOH) λmax (Δε) 202 (–11.12), 225 (+7.96), 240 (–3.56), 255 (+2.29), 272 (–2.72) nm; 1H and 13C NMR data, see Tables 1 and 2, Supporting Information S14–S19; HRESIMS m/z 528.2112 [M + Na]+ (calcd for C28H31N3O6Na, 528.2105).
10-O-ethylsclerotiamide (4): white powder, [α]20D –66 (c 0.02, MeOH); UV (MeOH) λmax (log ε) 317 (3.19), 242 (4.31), 198 (4.33) nm; CD (0.41 mM, MeOH) λmax (Δε) 199 (–10.70), 224 (+7.51), 240 (–7.64), 255 (+1.01), 272 (–2.91) nm; 1H and 13C NMR data, see Tables 1 and 2, Supporting Information S20–S25; HRESIMS m/z 514.2316 [M + Na]+ (calcd for C28H33N3O5Na, 514.2312).
10-O-ethylnotoamide R (5): white powder, [α]20D + 27 (c 0.04, MeOH); UV (MeOH) λmax (log ε) 305 (3.88), 239 (4.45), 212 (4.39), 186 (4.42) nm; CD (0.42 mM, MeOH) λmax (Δε) 203 (–16.25), 224 (+16.59), 247 (–1.77) nm; IR (CDCl3) νmax 2977, 1676, 1645, 1507 cm−1; 1H and 13C NMR data, see Tables 1 and 2, Supporting Information S26–S31; HRESIMS m/z 498.2332 [M + Na]+ (calcd for C28H33N3O4Na, 498.2363).
The crystallographic data for 1: (C26H31N3O5)2·3H2O, Mr = 985.12, monoclinic, space group P21 with a = 11.2882(11) Å, b = 14.2766(14) Å, c = 16.0232(15) Å, β = 101.3281(14)°, V = 2531.9(4) Å3, Z = 2, Dcal = 1.292 g/cm3, µ = 0.093 mm−1, and F(000) = 1052.
X-ray crystallographic analysis of 17-hydroxynotoamide D (1)
Experimental intensity data for (C26H31N3O5))2·3H2O were collected at T = 200(2) K on a BRUKER Kappa APEX2 diffractometer with graphite monochromated Mo Kα radiation (λ = 0.71073 Å). Intensity data were corrected for absorption using the multi-scan method. The structure was solved using direct methods and refined by least-squares calculation in anisotropic approximation for non-hydrogen atoms. The H atoms of the water molecules was located in an electron density map and its coordinates were freely refined with Uiso = 1.5Ueq(O). The rest hydrogen atoms were added at ideal positions and refined using a riding model. The asymmetric unit of the structure contains two molecules 1 and three molecules H2O. In one of non-equivalent molecules of 1 the C(31) carbon atom in proline ring is disordered equiprobably over two positions (Supporting Information, Figure S7). Data collection and editing, as well as refinement of unit cell parameters, were performed with the APEX2 program packages [23]. Structure solution and refinement were performed with the SHELXTL/PC program packages [24]. Complete crystallographic details are included in the Supporting Information, Figure S39. Selected bond lengths, bond angles, and hydrogen bonds are listed in Figure S39. Supplementary crystallographic data (accession number CCDC 1479246) can be obtained free of charge from the Cambridge Crystallographic Data Center via http://www.ccdc.cam.ac.uk/data_request/cif (or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB21EZ, U.K.; fax: + 44 1223 336 033 or email: deposit@ccdc.cam.uk).
Biological assay
Cell lines and culture conditions
The human prostate cancer cells lines 22Rv1, PC-3, and LNCaP, human embryonic kidney cell line HEK 293 T, and human fibroblast cell line MRC-9 were purchased from ATCC. Cell lines were cultured according to the manufacturer’s instructions in RPMI media (Invitrogen) with (for LNCaP) or without (for 22Rv1 and PC-3) 1 mM sodium pyruvate (Invitrogen); MRC-9 and HEK 293 T cells were cultured in DMEM medium (Invitrogen). The medium were supplemented with GlutamaxTM-I (Invitrogen, Paisley, UK) and contained 10% FBS (Invitrogen) and 1% penicillin/streptomycin (Invitrogen). The cells were continuously kept in culture for a maximum of three months and were routinely inspected microscopically for stable phenotype and regularly checked for contamination with mycoplasma. Cell line authentication was performed by DSMZ (Braunschweig, Germany) using highly polymorphic short tandem repeat loci [25].
In vitro MTT-based drug sensitivity assay
The in vitro cytotoxicity of individual substances was evaluated using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay, which was performed as previously described [26].
Colony formation assay
Colony formation assay was performed as described before with slight modifications [25]. The cells were treated with the drug for 48 h, then the cells were trypsinized, and the number of alive cells was counted with the trypan blue exclusion assay as described before [27]. 100 viable cells were plated into each well of 6-well plates in complete drug-free media (3 mL/well) and were incubated for 14 days. Then, the media was aspirated, surviving colonies were fixed with 100% MeOH, followed by washing with PBS, and air-dried at RT. Next, the cells were incubated with the Giemsa staining solution was for 25 min at RT, the staining solution was aspirated, wells were rinsed with dH2O, and air-dried. Number of cell colonies was counted with the naked eye.
References
Blunt JW, Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat Prod Rep. 2018;35:8–53.
Blunt JW, Copp BR, Keyzers RA, Munro MHG, Prinsep MR. Marine natural products. Nat Prod Rep. 2017;34:235–94.
Rateb ME, Ebel R. Secondary metabolites of fungi from marine habitats. Nat Prod Rep. 2011;28:290–344.
Wang Y, Wang L, Zhuang Y, Kong F, Zhang C, Zhu W. Phenolic polyketides from the co-cultivation of marine-derived Penicillium sp. WC-29-5 and Streptomyces fradiae 007. Mar Drugs. 2014;12:2079–88.
Meng LH, Liu Y, Li XM, Xu GM, Ji NY, Wang BG. Citrifelins A and B, citrinin adducts with a tetracyclic framework from cocultures of marine-derived isolates of Penicillium citrinum and Beauveria felina. J Nat Prod. 2015;78:2301–5.
Li C, Wang J, Luo C, Ding W, Cox DG. A new cyclopeptide with antifungal activity from the co-culture broth of two marine mangrove fungi. Nat Prod Res. 2014;28:616–21.
Smetanina OF, Yurchenko AN, Afiyatullov SS, Kalinovsky AI, Pushilin MA, Khudyakova YV, Slinkina NN, Ermakova SP, Yurchenko EA. Oxirapentyns B-Dproduced by a marine sediment-derived fungus Isaria felina (DC.) Fr. Phytochem Lett. 2012;5:165–9.
Yurchenko AN, Smetanina OF, Khudyakova YV, Kirichuk NN, Chiakina EL, Anisimov MM, Afiyatullov SS. New oxirapentyn E from marine isolate of the fungus Isaria felina. Chem Nat Comp. 2013;49:857–60.
Yurchenko AN, Smetanina OF, Kalinovsky AI, Pushilin MA, Glazunov VP, Khudyakova YV, Kirichuk NN, Ermakova SP, Dyshlovoy SA, Yurchenko EA, et al. Oxirapentyns F-K from the marine-sediment-derived fungus Isaria felina KMM 4639. J Nat Prod. 2014;77:1321–8.
Zhuravleva OI, Afiyatullov SS, Vishchuk OS, Denisenko VA, Slinkina NN, Smetanina OF. Decumbenone C, a new cytotoxic decaline derivative from the marine fungus Aspergillus sulphureus KMM 4640. Arch Pharm Res. 2012;35:1757–62.
Zhuravleva OI, Kirichuk NN, Denisenko VA, Dmitrenok PS, Yurchenko EA, Min′ko EM, Ivanets EV, Afiyatullov SS. New diorcinol J produced by co-cultivation of marine fungi Aspergillus sulphureus and Isaria felina. Chem Nat Comp. 2016;52:227–30.
Smetanina OF, Yurchenko AN, Ivanets EV, Kalinovsky AI, Khudyakova YV, Dyshlovoy SA, Von Amsberg G, Yurchenko EA, Afiyatullov SS. Unique prostate cancer-toxic polyketides from marine sediment-derived fungus Isaria felina. J Antibiot. 2017;70:856–8.
Kato H, Yoshida T, Tokue T, Nojiri Y, Hirota H, Ohta T, Williams RM, Tsukamoto S, Notoamides A-D.Prenylated indole alkaloids isolated from a marine-derived fungus, Aspergillus sp. Angew Chem Int Ed. 2007;46:2254–6.
Oki M, Iwamura H. Intramolecular interaction between hydroxyl group and π-electrons. VII. limitation of the interaction by chain length in 2-(ω-alkenyl)-phenols and 2-(ω-phenylalkyl)-phenols. Bull Chem Soc Jpn. 1960;33:681–4.
Berdyshev DV, Glazunov VP, Novikov VL. Conformational analysis and interpretation of ν(OH) band in the IR spectrum of o-vinylphenol: A DFT study. Russ Chem Bull Int Ed. 2008;57:510–9.
Tsukamoto S, Kawabata T, Kato H, Greshock T, Hirota H, Ohta T, Williams RM. Isolation of antipodal (-)-versicolamide B and notoamides L-N from a marine-derived Aspergillus sp. Org Lett. 2009;11:1297–1300.
Tsukamoto S, Umaoka H, Yoshikawa K, Ikeda T, Hirota H. Notoamide O. a structurally unprecedented prenylated indole alkaloid, and notoamides P-R from a marine-derived fungus Aspergillus sp. J Nat Prod. 2010;73:1438–40.
Chen M, Shao CL, Fu XM, Xu RF, Zheng JJ, Zhao DL. She ZGM, Wang CY. Bioactive indole alkaloids and phenyl ether derivatives from a marine-derived Aspergillus sp. fungus. J Nat Prod. 2013;76:547–53.
Whyte AC, Gloer JB, Wicklow DT, Dowd PF. Sclerotiamide: a new member of the paraherquamide class with potent antiinsectan activity from the sclerotia of Aspergillus sclerotiorum. J Nat Prod. 1996;59:1093–5.
Tsukamoto S, Kato H, Samizo M, Onuki H, Nojiri Y, Hirota H, Ohta T. Notoamides F-K, prenylated indole alkaloids isolated from a marine-derived Aspergillus sp. J Nat Prod. 2008;71:2064–7.
Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, Gao AC. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res. 2014;20:3198–210.
Kato H, Nakahara T, Sugimoto K, Matsuo K, Kagiyama I, Frisvad JC, Sherman DH, Williams RM, Tsukamoto S. Isolation of notoamide S and enantiomeric 6-epi-stephacidin A from the fungus Aspergillus amoenus: biogenetic implications. Org Lett. 2015;17:700–3.
Bruker. APEX2, Bruker AXS Inc. USA: Madison, Wisconsin; 2012.
Sheldrick GM. Crystal structure refinement with SHELXL. Acta Cryst. 2015;A71:3–8.
Dyshlovoy SA, Menchinskaya ES, Venz S, Rast S, Amann K, Hauschild J, Otte K, Kalinin VI, Silchenko AS, Avilov SA, Alsdorf W, Madanchi R, Bokemeyer C, Schumacher U, Walther R, Aminin DL, Fedorov SN, Shubina LK, Stonik VA, Balabanov S, Honecker F, Amsberg G. The marine triterpene glycoside frondoside A exhibits activity in vitro and in vivo in prostate cancer. Int J Cancer. 2016;138:2450–65.
Dyshlovoy SA, Venz S, Shubina LK, Fedorov SN, Walther R, Jacobsen C, Stonik VA, Bokemeyer C, Balabanov S, Honecker F. Activity of aaptamine and two derivatives, demethyloxyaaptamine and isoaaptamine, in cisplatin-resistant germ cell cancer. J Proteom. 2014;96:223–39.
Dyshlovoy SA, Hauschild J, Amann K, Tabakmakher KM, Venz S, Walther R, Guzii AG, Makarieva TN, Shubina LK, Fedorov SN, Stonik VA, Bokemeyer C, Balabanov S, Honecker F, Amsberg G. Marine alkaloid monanchocidin A overcomes drug resistance by induction of autophagy and lysosomal membrane permeabilization. Oncotarget. 2015;6:17328–41.
Acknowledgements
The study was supported by the Ministry of Science and High Education of Russian Federation to support the bioresource collections. The authors are grateful to the Far Eastern Center of Structural Research for performing the X-ray investigation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Afiyatullov, S.S., Zhuravleva, O.I., Antonov, A.S. et al. Prenylated indole alkaloids from co-culture of marine-derived fungi Aspergillus sulphureus and Isaria felina. J Antibiot 71, 846–853 (2018). https://doi.org/10.1038/s41429-018-0072-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-018-0072-9
This article is cited by
-
Recent advances in microbial co-culture for production of value-added compounds
3 Biotech (2022)
-
Co-culture: stimulate the metabolic potential and explore the molecular diversity of natural products from microorganisms
Marine Life Science & Technology (2021)
-
Marine-derived fungi as a source of bioactive indole alkaloids with diversified structures
Marine Life Science & Technology (2021)
-
Advances in microbial culturing conditions to activate silent biosynthetic gene clusters for novel metabolite production
Journal of Industrial Microbiology and Biotechnology (2019)