Abstract
A novel endophytic actinobacterium, designated strain RL12-1ST, was isolated from surface-sterilized leaves of Oryza sativa L. collected from Roi Et province, Thailand. Taxonomic position of strain RL12-1ST was studied using the polyphasic approach. Phylogenetic analyses base on the 16S rRNA gene sequences showed that strain RL12-1ST belonged to the genus Quadrisphaera and was closely related to Quadrisphaera granulorum AG019T (99.0%). The colony of strain RL12-1ST was vivid orange. Growth occurred at a temperature between 17–37 °C and pH between 6.0–8.0. The strain contained meso-diaminopimelic acid and galactose, glucose, mannose, ribose and rhamnose in whole-cell hydrolysates. The predominant menaquinone was MK-8(H2). The major fatty acids were C16:1ω7c/C16:1ω6c, C16:0 and C18:1ω7c. The polar lipids of the strain were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and phosphatidylinositol mannosides. The DNA G + C content of genomic DNA was 76.1 mol%. Based on comparative analysis of physiological, biochemical and chemotaxonomic data, including DNA–DNA hybridization, strain RL12-1ST represents a novel species of the genus Quadrisphaera, for which the name Quadrisphaera oryzae sp. nov. is proposed. The type strain is RL12-1ST ( = TBRC 8486T = NBRC 113447T).
Similar content being viewed by others
Introduction
The genus Quadrisphaera was first proposed by Maszenan et al. [1]. and belong to the family Kineosporiaceae, which accommodates genera Angustibacter, Kineococcus, Kineosporia, Pseudokineococcus, Quadrisphaera and Thalassiella [2]. Currently, the genus Quadrisphaera contained only one validly published species which is Quadrisphaera granulorum AG019T and is the type species [3]. The strain was isolated from aerobic granules in a fed-batch activated sludge reactor. Member of the genus is Gram-stain-positive cocci, non-spore-forming, non-motile, occurring in tetrad arrangements. The predominant of menaquinone is MK-8(H2) [4]..In this study, a novel Quadrisphaera species with designated strain RL12-1ST, isolated from surface-sterilized leaves of rice plant (Oryza sativa L.), was described using polyphasic taxonomy.
Material and methods
Isolation and culture condition
During the investigation of actinomycetes diversity in rice plants (Oryza sativa L.) from northeast of Thailand, strain RL12-1ST was isolated from surface-sterilized rice leaves collected from Roi Et province, Thailand. The leaves samples were surface-sterilized as previously described by Himaman et al. [5]. Briefly, leaves samples were cut in segments and subjected to a surface sterilization by soaking in 0.1% Tween 20 for 5 min, followed by a solution of 1.0% NaOCl for 10 min, 2.5% Na2S2O3 for 10 min and 75% ethanol for 5 min. Samples were then rinsed with sterile distilled water thrice to remove surface sterilization agents. The surface sterile rice leaves were then soaked in 10% NaHCO3 for 10 min before crushed with a sterile glass rod and diluted with 0.85% NaCl. An aliquot of 100 µl of tissues suspension was spread on starch casein (SC) agar (1% soluble starch, 0.03% casein, 0.2% KNO3, 0.2% NaCl, 0.2% KH2PO4, 0.002% CaCO3, 0.005% MgSO4·7H2O, 0.001% FeSO4·7H2O and 1.8% agar; pH 7.2) supplemented with nystatin (50 µg ml−1), ketoconazole (100 µg ml−1) and nalidixic acid (25 µg ml−1). Plates were incubated at 28 °C for 14 days. A single vivid orange colony of strain RL12-1ST was isolated, purified and maintained on International Streptomyces Project medium 2 (ISP2) [6] slants at room temperature and as 20% (v/v) glycerol suspensions at –20 °C. The type strain of Quadrisphaera granulorum JCM 16010T was used in this study for comparison purposes.
Morphological and physiological characteristics
Cultural and morphological characteristics of strain RL12-1ST were determined after incubation for 7 days at 28 oC on various media including ISP2–ISP7 agar [6] and glucose yeast extract agar (GYE) [7]. The colour of colonies and soluble pigments were determined by comparing with the standard names of the NBS/ISCC Color System [8]. Gram staining was carried out by using the standard Gram reaction method. Cell morphological characteristics of strain RL12-1ST were observed by light microscopy (Nikon ECLIPSE E100, Tokyo, Japan) fit with long working distance objective lens, and scanning electron microscopy (Quanta 450 FEI). Catalase activity was detected by the production of bubbles after adding a few drops of 3% (v/v) hydrogen peroxide solution onto the cells. Oxidase activity was examined from the oxidation of 1% tetramethyl-p-phenylenediamine dihydrochloride solution. The temperature for growth was determined at different temperature (17, 20, 25, 28, 30, 33, 35, 37, 40, 42 and 45 °C) on ISP2 agar slant by using a temperature gradient incubator model TN-3, Toyo Kagaku Sangyo (Tokyo, Japan). The effects of NaCl concentrations (0–10%, w/v at intervals of 1%) on growth were determined on ISP2 agar. The ability to grow in different pH values (pH 3.0–10.0, at intervals of 1.0 pH units) was studied in ISP2 broth. The medium was adjusted to the appropriate pH with the buffer system: 0.1 M citric acid/0.1 M sodium citrate (pH 3.0–5.0); 0.1 M KH2PO4/0.1 M NaOH (pH 6.0–8.0); 0.1 M NaHCO3/0.1 M Na2CO3 (pH 9.0–10.0) [5]. Carbon source utilization was examined using ISP9 agar as a basal medium, supplemented with 1% final concentration of carbon sources [6]. Enzyme activity profiles were tested using API® ZYM test kit (bioMérieux) according to the manufacturer’s instructions. Utilization of nitrogen sources, urease activity, degradation activity and lysozyme resistance were examined using the media according to Gordon et al. [7] and Williams et al. [9]. The hydrogen sulfide production was examined using lead acetate stripes as detector according to the method described by Küster and Williams [10].
Chemotaxonomic analysis
Biomass for chemotaxonomic studies was obtained by growing the strains in GYE broth and incubated at 28 °C on a shaking incubator maintained at 150 rpm for 4 days. Cells were harvested by centrifugation and washed three times with sterile distilled water before freeze-drying. The isomers of diaminopimelic acid and sugar analysis in the whole-organism hydrolysates were determined by thin-layer chromatography (TLC) according to the standard procedures described by Hasegawa et al. [11] and Staneck and Robert [12]. Polar lipids were extracted from freeze-dried cell and examined by two-dimensional TLC as previously described by Minnikin et al. [13]. The N-acyl type of muramic acid in cell wall peptidoglycan was analysed according to the method of Uchida et al. [14]. Menaquinones were extracted and purified using the method of Collins et al. [15] and isoprene units were subsequently analysed by LC/MS (JMS-T100LP; JEOL) with a CAPCELL PAK C18 UG120 column (Shiseido) using methanol: 2-propanol (7: 3). Mycolic acids were detected by TLC according to the method of Tomiyasu [16]. The fatty acid analysis was determined using GLC according to the instructions of the Sherlock Microbial Identification System (Microbial ID; MIDI, Version 6.2B) using the RTSBA6 database [17] for both RL12-1ST and Q. granulorum JCM 16010T. The analysis was performed at Thailand Institute of Scientific and Technological Research (TISTR). The G + C content of the genomic DNA was determined by HPLC according to the method of Tamaoka and Komagata [18]. Levels of DNA–DNA hybridization between strain RL12-1ST and member of the closely related species of genus Quadrisphaera were determined fluorometrically by the method of Ezaki et al. [19].
16S rRNA gene sequencing and phylogenetic analysis
The genomic DNA of strain RL12-1ST was extracted and purified according to Kieser et al. [20]. The 16S rRNA gene was amplified using primers 1F (5′ ˗TCACGGAGAGTTTGATCCTG˗3′) and 1530R (5′˗AAGGAGATCCAGCCGCA˗3′), under the conditions as described by Mingma et al. [21]. The PCR products were purified using a FavorPrepTM Gel/PCR Purification Kit (FAVORGEN®, BIOTECH CORP., Taiwan) following the manufacturer’s protocol. Sequencing of the 16S rRNA gene was performed using the service of Macrogen (Seoul, South Korea) with primers 1F, 1530R, Mg4F (5′˗AATTCCTGGTGTAGCGGT˗3′) and 782R (5′˗ACCAGGGTATCTAATCCTGT˗3′). The obtained 16S rRNA gene sequence (1,472 nt) was aligned with a member of the genus Quadrisphaera and sequences of representative genera in family Kineosporiaceae, which sequences downloaded from EzBioCloud server (https://www.ezbiocloud.net/) [22]. Evolutionary trees were inferred using maximum-parsimony [23], maximum-likelihood [24] and neighbour-joining [25] tree-making algorithms drawn from the MEGA 6 package [26]. The topology of the phylogenetic tree was evaluated by carrying out a bootstrap analysis on 1,000 resampled datasets [27].
Nucleotide sequence accession number
The 16S rRNA gene sequence of strain RL12-1ST was deposited to the DDBJ under accession number LC405931.
Results and discussion
An almost complete 16S rRNA gene sequence of strain RL12-1ST (1,472 nt) was used for phylogenetic analysis. The result showed that the strain RL12-1ST was a member of the genus Quadrisphaera. The sequence similarity indicated that the most closely related strain was Q. granulorum AG019T (99.0%). It is evident from the neighbour-joining tree that strain RL12-1ST formed a distinct cluster with the type strain of Q. granulorum AG019T, that was supported by a relative high bootstrap value of 100% (Fig. 1) and by all of the tree-making algorithms of maximum-parsimony and maximum-likelihood. The taxonomic position of the strain RL12-1ST was supported by DNA–DNA relatedness data. Strain RL12-1ST showed DNA–DNA relatedness values of 33.7–40.0% to Q. granulorum JCM 16010T. These values are well below the delineating 70% cutoff point for species identification [28]. The genomic DNA G + C content of strain RL12-1ST was 76.1 mol%.
Neighbour-joining phylogenetic tree based on almost-complete 16S rRNA gene sequences, showing the relationship of strain RL12-1ST and its closest phylogenetic relatives. Asterisks indicate clades that were conserved using the maximum-parsimony and maximum-likelihood tree. Only values above 50% (percentages of 1,000 replications) are given. Bar, 0.01 nucleotide substitutions per position
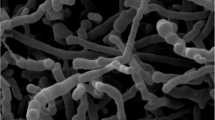
Strain RL12-1ST was aerobic, non-spore-forming, Gram-stain-positive actinobacterium and non-motile. The cells were cocci, 1.2 μm in diameter, occurred in tetrads (Fig. 2a). In young cultures, the strain formed ovoid and slender bud shaped (Fig. 2b) in both solid and liquid media. Morphological characteristics on agar media revealed that the colonies were circular, convex and vivid orange in colour. The strain exhibited good growth on ISP2 and GYE agar, moderate on ISP6 but poor growth on ISP3, ISP4, ISP5 and ISP7 after incubation at 28 °C for 7 days. Melanin and diffusible pigments were not observed on all tested media. The cultural characteristics of strain RL12-1ST on various media are shown in Supplementary Table S1. The strain RL12-1ST showed oxidase negative and catalase positive. The temperature range for growth was between 17–37 °C with optimum growth at 25–33 °C. No growth was observed at or above 40 °C. The strain was able to grow at pH 6.0–8.0, with optimal growth at pH 6.0–7.0. The strain grown in NaCl concentrations of 0–5% (w/v). No growth was observed in the presence of 6% NaCl (w/v) or higher.
Whole–cell hydrolysates of strain RL12-1ST contained meso–diaminopimelic acid, galactose, glucose, mannose, rhamnose and ribose. The menaquinones were MK-8(H2) (95.3%), MK-9(H2) (3.0%) and MK-8(H4) (1.7%) and cells lacked mycolic acid. The polar lipids of the strain were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, unknown polar lipids and unknown phospholipid (Supplementary Figure S1). The major fatty acids were C16:1ω7c/C16:1ω6c, C16:0 and C18:1ω7c. The cellular fatty acid composition of strain RL12-1ST and Q. granulorum JCM 16010T, the closely related species, can be found in Supplementary Table S2. Cell wall acyl type was N–acetylated.
In addition, strain RL12-1ST could be differentiated from Q. granulorum JCM 16010T based on physiological and biochemical properties given in Table 1. Strain RL12-1ST was able to utilize L-asparagine and L-histidine as sole nitrogen sources for growth, whereas Q. granulorum JCM 16010T could not. Strain RL12-1ST showed good growth on GYE agar, but Q. granulorum JCM 16010T showed moderate growth. Casein, urea and xanthine were degraded by strain RL12-1ST, but not by Q. granulorum JCM 16010T. Furthermore, Q. granulorum JCM 16010T degraded L-tyrosine but strain RL12-1ST did not. H2S production was weakly positive in strain RL12-1ST, whereas Q. granulorum JCM 16010T was negative. Strain RL12-1ST utilized D(–)fructose, D(–)mannitol, D(–)sorbitol, D( + )galactose, D( + )mannose, D( + )trehalose, maltose and raffinose as a sole carbon sources, but Q. granulorum JCM 16010T showed negative results. Based on the phylogenetic analysis, phenotypic characteristics and DNA–DNA relatedness data, it is concluded that strain RL12-1ST represents a novel species of the genus Quadrisphaera, for which name Quadrisphaera oryzae sp. nov. is proposed.
Description of Quadrisphaera oryzae sp. nov
Quadrisphaera oryzae (o.rý zae. L. gen. n. oryzae of rice, pertaining to the isolation of the type strain from leaves of rice plant).
Cells are aerobic, Gram-stain-positive cocci, non-spore-forming, occurring in tetrads arrangement. Colonies are circular, convex and vivid orange in colour. Strain RL12-1ST are catalase positive but oxidase negative. Grow well on ISP2 and GYE agar. Moderate growth occurs on ISP6 agar but poor growth on ISP3, ISP4, ISP5 and ISP7 media. Melanin pigments are not detected. Nitrate reduction is negative. Hydrogen sulfide production is observed. Growth occurs between 17–37 °C with optimum growth at 25–33 °C. No growth at or above 40 °C. Grow at pH 6.0–8.0, with optimal growth at pH 6.0–7.0. Good growth occurs in ISP2 containing NaCl concentrations 0–2% (w/v), moderate growth at 3% and poor growth at 4–5%. No growth in the presence of 6% NaCl (w/v). Aesculin, arbutin, casein, starch, Tween 80, urea and xanthine are degraded. Adenine, cellulose, gelatin, guanine, hypoxanthine, L-tyrosine, Tween 20 and xylan are not degraded. L-Asparagine, L-histidine and KNO3 are used as a sole nitrogen sources. Utilizes D(–)glucose, D(–)fructose, D(–)lactose, D(–)mannitol, D(–)ribose, D(–)sorbitol, D( + )cellobiose, D( + )galactose, D( + )mannose, D( + )trehalose, D( + )xylose, L( + )arabinose, maltose, myo-inositol and raffinose as a sole carbon sources. L( + )Rhamnose, sucrose, xylitol, sodium citrate and sodium propionate are not utilized. Alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase, acid phosphatase, naphthol–AS–BI–phosphohydrolase, α–galactosidase, β–galactosidase, α–glucosidase, β–glucuronidase and α–fucosidase are positive. Lipase (C14), trypsin, α–chymotrypsin, β–glucosidase, N–acetyl–β–glucosaminidase and α–mannosidase are negative. Not resistant to 0.005% lysozyme. Whole-cell hydrolysates contains meso-diaminopimelic acid. Mycolic acids are not detected. The menaquinones are MK-8(H2), MK-9(H2) and MK-8(H4). Whole-cell sugars are galactose, glucose, mannose, rhamnose and ribose. Polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, two unknown polar lipids and an unknown phospholipid. The major fatty acids are C16:1ω7c/C16:1ω6c, C16:0 and C18:1ω7c. Cell wall acyl types are N–acetylated. The DNA G + C content of type strain is 76.1 mol%. The type strain is RL12-1ST ( = TBRC 8486T = NBRC 113447T), isolated from the surface–sterilized leaves of rice plant (Oryza sativa L.), collected from Roi Et province, Thailand.
References
Maszenan AM, Tay J-H, Schumann P, Jiang H-L, Tay ST-L. Quadrisphaera granulorum gen. nov., sp. nov., a Gram-positive polyphosphate-accumulating coccus in tetrads or aggregates isolated from aerobic granules. Int J Syst Evol Microbiol. 2005;55:1771–7.
Oren A, Garrity GM. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2017;67:1–3.
Parte AC. LPSN – List of Prokaryotic Names with Standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol. 2018;68:1825–9.
Norman P, Benson, DR. Genus III. Quadrisphera. In: Goodfellow M, et al. editors. Bergey’s Manual of Systematic Bacteriology, 2nd edn, Vol. 5, Springer: New York, 2012. p. 567–9.
Himaman W, Thamchaipenet A, Pathom-aree W, Duangmal K. Actinomycetes from Eucalyptus and their biological activities for controlling Eucalyptus leaf and shoot blight. Microbiol Res. 2016;188:42–52.
Shirling ET, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Evol Microbiol. 1966;16:313–40.
Gordon RE, Barnett DA, Handerhan J, Pang CH-N. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol. 1974;24:54–63.
Mundie D. The NBS/ISCC Color System/David A. Mundie Pittsburgh, PA: Polymath Systems 1995; 535–6.
Williams ST, et al. Numerical classification of Streptomyces and related genera. Microbiology. 1983;129:1743–813.
Küster E, Williams ST. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–9.
Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–22.
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–31.
Minnikin D, Patel P, Alshamaony L, Goodfellow M. Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Evol Microbiol. 1977;27:104–17.
Uchida K, Kudo T, Suzuki K-i, Nakase T. A new rapid method of glycolate test by diethyl ether extraction, which is applicable to a small amount of bacterial cells of less than one milligram. J Gen Appl Microbiol. 1999;45:49–56.
Collins M, Pirouz T, Goodfellow M, Minnikin D. Distribution of menaquinones in actinomycetes and corynebacteria. Microbiology. 1977;100:221–30.
Tomiyasu I. Mycolic acid composition and thermally adaptative changes in Nocardia asteroides. J Bacteriol. 1982;151:828–37.
Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101.(MIDI Inc., Newark, DE, USA, 1990)
Tamaoka J, Komagata K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett. 1984;25:125–8.
Ezaki T, Hashimoto Y, Yabuuchi E. Fluorometric deoxyribonucleic acid deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Evol Microbiol. 1989;39:224–9.
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000.
Mingma R, Pathom-aree W, Trakulnaleamsai S, Thamchaipenet A, Duangmal K. Isolation of rhizospheric and roots endophytic actinomycetes from Leguminosae plant and their activities to inhibit soybean pathogen, Xanthomonas campestris pv. glycine. World J Microbiol Biotechnol. 2014;30:271–80.
Yoon S-H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–7.
Fitch WM. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst Zool. 1971;20:406–16.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA 6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Wayne L, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–4.
Acknowledgements
SM was awarded a Ph.D. scholarship by the Royal Golden Jubilee of the Thailand Research Fund (RGJ-TRF) (Grant No. PHD02022558). We would like to thank Kasetsart University Research and Development Institute (KURDI) for research funding (2561:3.1–35.61).
The DDBJ accession number for the 16S rRNA gene sequence of strain RL12-1ST is LC405931 (1,472 nt).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Muangham, S., Lipun, K., Matsumoto, A. et al. Quadrisphaera oryzae sp. nov., an endophytic actinomycete isolated from leaves of rice plant (Oryza sativa L.). J Antibiot 72, 93–98 (2019). https://doi.org/10.1038/s41429-018-0112-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-018-0112-5
This article is cited by
-
Microbispora oryzae sp. nov., isolated from leaves of rice plant (Oryza sativa L.)
The Journal of Antibiotics (2021)