Abstract
A polyphasic approach was used to identify the novel actinomycete, strain 10-20SHSuT, isolated from the rhizosphere of the mangrove associated plants Suaeda maritima collected from Phetchaburi Province, Thailand. Phylogenetic analysis based on 16S rRNA gene sequences indicated that the organism belonged to the phylogenetic cluster of the genus Nonomuraea and was most closely related to Nonomuraea soli YIM 120770T (98.1% sequence similarity), Nonomuraea endophytica YIM 65601T (97.3%) and Nonomuraea candida HMC10T (97.3%). The strain formed an extensively branched substrate and aerial mycelia. The whole-cell hydrolysates contained meso-diaminopimelic acid as the diagnostic diamino acid, with galactose, glucose, madurose, mannose and ribose as the whole-cell sugars. The polar lipids were diphosphatidylglycerol, hydroxy-phosphatidylethanolamine, hydroxy-phosphatidylmonomethylethanolamine, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol mannosides, phosphatidylinositol, phosphotidylmethylethanolamine, two unidentified sugar containing phosphoaminolipids and an unidentified phospholipid. MK-9(H4) was a major menaquinone of the organism. The predominant cellular fatty acids were iso-C16:0, C17:0 and 10-methyl-C17:0. The G + C content of the genomic DNA was 71.9 mol%. On the basis of phenotypic characteristics, DNA–DNA relatedness and phylogenetic distinctiveness, strain 10-20SHSuT represents a novel species of the genus Nonomuraea, for which the name Nonomuraea suaedae sp. nov. is proposed. The type strain is 10-20SHSuT (=TBRC 8487T =NBRC 113448T).
Similar content being viewed by others
Introduction
The genus Nonomuraea (corrig.) was first proposed by Zhang et al. [1], belongs to the family Streptosporangiaceae within the class Actinobacteria. The family encompasses 15 genera, namely, Streptosporangium (type genus), Acrocarpospora, Herbidospora, Microbispora, Microtetraspora, Nonomuraea, Planobispora, Planomonospora, Planotetraspora, Sphaerimonospora, Sphaerisporangium, Thermoactinospora, Thermocatellispora, Thermopolyspora and Thermostaphylospora [2]. Although members of the family Streptosporangiaceae are chemically homogeneous, despite some differences, members of the genus Nonomuraea are readily distinguished from members of related genera based on 16S rRNA gene sequence [3]. Furthermore, members of this genus are filamentous Gram-stain positive bacteria which generally form extensively branched substrate and aerial mycelia with either hooked, spiral or straight chains of spores observed on the aerial mycelium. The spore surfaces can be folded, irregular, smooth or warty [4]. Additionally, the sequence similarities within the genus Nonomuraea ranged from 93.9 to 99.8% [5]. The herbaceous plant sample, Sueda maritima is a widely distributed halophyte of mangrove associated plants that flourish in saline moist soil [6]. In Thailand, this plant is distributed along the coastal forest of the Gulf of Thailand [7]. In the present study, taxonomic position of strain 10-20SHSuT, isolated from rhizosphere soil of Sueda maritima was determined using a polyphasic approach and it is proposed as representing a novel species of the genus Nonomuraea.
Materials and methods
Bacterial strains and isolation
Strain 10-20SHSuT was isolated from the rhizosphere of Suaeda maritima (L.) Dumort, collected from Phetchaburi Province, Thailand. Collected plant roots were suspended in sterile distilled water and briefly vortexed. Rhizosphere soil obtained from the suspension was serially diluted and plated on humic acid-vitamin agar [8] supplemented with nalidixic acid (25 µg ml–1) and nystatin (50 µg ml–1) as the selective medium. After the plates were incubated at 30 °C for 21 days, the strain was transferred and purified on glucose yeast extract (GYE) agar (containing glucose 1.0%, yeast extract 1.0% and agar 1.5%, w/v). The pure culture of strain 10-20SHSuT was maintained on GYE agar slants for further studies. Mycelia and spores were kept in glycerol solution (20%, v/v) at –20 °C for long-term preservation. Nonomuraea soli JCM 17347T, Nonomuraea endophytica JCM 31210T and Nonomuraea candida JCM 15928T were used for morphological, physiological, chemotaxonomic and molecular taxonomic studies.
Morphological, physiological, and biochemical tests
The cultural and growth characteristics of strain 10-20SHSuT were examined on International Streptomyces Project (ISP) [9] media 2, 3, 4, 5, 6 and 7 and GYE agar, after incubation at 28 °C for up to 21 days. The colour of substrate and aerial mycelia were determined by comparing with the Colour Harmony Manual [10]. Mycelium formation was observed under scanning electron microscopy (Quanta 450 FEI) after the strain was cultivated on ISP 2 agar at 28 °C for 10 days. The temperature range for growth was determined on ISP 2 using a temperature gradient incubator over the temperature range 14–45 °C for 14 days. Growth at different pH (pH 4.0–9.0 at intervals of 1.0 pH units), the medium was adjusted to the appropriate pH with the buffer system: 0.1 M citric acid/0.1 M sodium citrate (pH 4–5); 0.1 M KH2PO4/0.1 M NaOH (pH 6–8); 0.1 M NaHCO3/0.1 M Na2CO3 (pH 9–10). NaCl concentration (0–10% at intervals of 1%, w/v) were examined on ISP 2 after incubation at 28 °C for 14 days. Utilization of carbohydrates as sole carbon source at a final concentration of 1% (w/v) was investigated on ISP 9 [9]. The utilization of (w/v) nitrogen sources (1.0%), lysozyme resistance (0.005%) and hydrolysis of adenine (0.4%), aesculin (0.1%), arbutin (0.1%), cellulose (1.0%), casein (skimmed milk, 5.0%), gelatin (0.4%), guanine (0.4%), hypoxanthine (0.4%), starch (1.0%), Tween 20 (1.0%, v/v), Tween 80 (1.0%, v/v), l-tyrosine (0.4%), urea (1.8%), xanthine (0.4%) and xylan (0.4%) were determined using various media as described by Gordon et al. [11] and Williams et al. [12]. Enzyme activities were determined using the API ZYM system (bioMérieux) following the instructions of manufacturer. Catalase and oxidase activities were determined with 3% (v/v) hydrogen peroxide solution and 1% (w/v) tetramethyl-p-phenylenediamine dihydrochloride solution, respectively. Nitrate reduction and H2S production were also studied following standard methods [13].
Chemotaxonomy
Freeze-dried cell for chemotaxonomic studies was prepared by growing the strain in shaking flasks of GYE broth at 28 °C for 3 days. Cultured cells were harvested by centrifugation, and the pellet was washed thrice with sterile distilled water prior to freeze drying. The isomer forms of diaminopimelic acid were determined by the methods of Becker et al. [14] and Hasegawa et al. [15]. Whole-cell sugars and the presence of mycolic acids were determined using TLC method [16, 17]. Polar lipids were extracted from freeze-dried cells and examined by two-dimensional TLC according to the procedure developed by Minnikin et al. [18]. Cellular menaquinones were extracted from freeze-dried biomass using the procedure of Collins et al. [19]. The cellular fatty acids were determined by using the Sherlock Microbial Identification System (version 6.2B; MIDI database: RTSBA6) according to the method of Sasser [20]. The analysis was performed at Thailand Institute of Scientific and Technological Research.
Molecular analysis
Genomic DNA of strain 10-20SHSuT was extracted and purified according to Kieser et al. [21]. The 16S rRNA gene was PCR amplified from genomic DNA using primers 1F (5′-TCACGGAGAGTTTGATCCTG-3′) and 1530R (5′-AAGGAGATCCAGCCGCA-3′), under the conditions as described by Mingma et al. [22]. The sequencing of the PCR product (by Macrogen, Korea) was performed using primers 1F, 1530R, Mg4F (5′-AATTCCTGGTGTAGCGGT-3′) and 782R (5′-ACCAGGGTATCTAATCCTGT-3′). The nearly complete 16S rRNA gene sequence of strain 10-20SHSuT (1447 nt) was compared to sequences of type strains in GenBank [23] and EzBioCloud [24] databases. Evolutionary trees were inferred with maximum-parsimony [25], maximum-likelihood [26] and neighbour-joining [27] tree-making algorithms in MEGA7 software package [28]. The resultant tree topologies were evaluated using a bootstrap analysis [29] on 1000 resampled datasets. The DNA G + C content was determined by HPLC according to the method of Tamaoka and Komagata [30]. DNA–DNA relatedness among strain 10-20SHSuT and closely related species of the genus Nonomuraea was investigated using the method as reported by Ezaki et al. [31].
Nucleotide sequence accession number
The GenBank accession number for the 16S rRNA gene sequence of strain 10-20SHSuT is MG757745.
Results and discussion
Morphological, cultural, and physiological characteristics
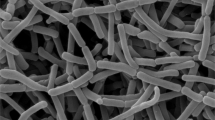
The morphological characteristics of strain 10-20SHSuT were consistent with these of members of the genus Nonomuraea. Strain 10-20SHSuT exhibited good growth on ISP 2 and GYE agar, moderate growth on ISP 3, ISP 4, ISP 5 and ISP 7 but poor growth on ISP 6 agar. The aerial mycelium appeared orange on ISP 2, ISP 3, ISP 4, ISP 6 and GYE agar; white on ISP 5 and ISP 7 agar. Brown soluble pigment was produced on ISP 2, ISP 3, ISP 4 and GYE agar but no soluble pigment was observed on ISP 5, ISP 6 and ISP 7 agar (Table S1). The strain formed an extensively branched substrate and aerial mycelia that formed straight or flexuous spore chains, which had bearing more than 10 spores. Sporangia were not found (Fig. 1). Growth occurred at 15–43 °C (optimum 27–38 °C), pH 5.0–9.0 (optimum pH 7.0–8.0) and in the presence of 0–3% (w/v) NaCl. Other details of the phenotypic characteristics are given in the species description and Table 1.
Chemotaxonomic characteristics
Strain 10-20SHSuT contained meso-diaminopimelic acid in the cell-wall diamino acid. Whole-cell hydrolysates contained madurose, galactose, glucose, mannose and ribose. Mycolic acids were absent. Polar lipids were diphosphatidylglycerol, hydroxy-phosphatidylethanolamine, hydroxy-phosphatidylmonomethylethanolamine, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol mannosides, phosphatidylinositol, phosphotidylmethylethanolamine, two unidentified sugar containing phosphoaminolipids and an unidentified phospholipid (Fig. S1). The predominant menaquinones were MK-9(H4) (59.9%) and MK-9(H2) (32.8%). Minor menaquinone was MK-9(H6) (4.7%). Major cellular fatty acids were iso-C16:0 (19.4%), 10-methyl-C17:0 (14.8%) and C17:0 (14.5%) and minor fatty acids were C16:0 (7.8%), iso-C15:0 (5.6%), iso-C14:0 (5.1%) and C14:0 (5.1%) (Table S2). The chemotaxonomic characteristics of strain 10-20SHSuT are typical for the genus Nonomuraea [1].
Molecular analysis
The pairwise 16S rRNA gene sequence similarity of strain 10-20SHSuT revealed that the similarity ranged from 98.1 to 94.4% with sequences of the type strains of Nonomuraea species with validly published names. The strain 10-20SHSuT showed the highest 16S rRNA gene sequence pairwise similarity with N. soli YIM 120770T (98.1%), N. endophytica YIM 65601T (97.3%) and N. candida HMC10T (97.3%). Furthermore, the phylogenetic tree based on neighbour-joining method of the 16S rRNA gene sequences showed that strain 10-20SHSuT formed a monophyletic clade with N. soli YIM 120770T (Fig. 2), wherein the branching was also recovered from phylogenetic trees based on maximum-likelihood and maximum-parsimony methods (Figs. S2 and S3). Phylogenetic tree with all members of the genus Nonomuraea is shown in Fig. S4. The genomic DNA G + C content of strain 10-20SHSuT was 71.9 mol%, within the range of 64–74 mol% of Nonomuraea species [3, 5]. Strain 10-20SHSuT also showed a DNA–DNA relatedness value of 55.9–59.3% to N. soli YIM 120770T, which was below the recommended cut-off point of 70% for species delineation [32]. Therefore, the genotypic data showed that strain 10-20SHSuT belongs to the genus Nonomuraea and can be distinguished from other species within the genus.
Neighbour-joining phylogenetic tree based on nearly complete 16S rRNA gene sequences showing the relationship between strain 10-20SHSuT and members of the genus Nonomuraea. Thermopolyspora flexuosa DSM 43186T was used as outgroup. Asterisks indicate that the corresponding branches were also recovered in both maximum-likelihood and maximum-parsimony phylogenetic trees. All positions containing gaps and missing data were eliminated. There were a total of 1232 positions in the final dataset. Bootstrap values > 50% (based on 1000 replications) are shown at branch points. Bar, 0.005 substitutions per nucleotide position
On the basis of the phenotypic, chemotaxonomic and phylogenetic characteristics presented, it is evident that strain 10-20SHSuT can be differentiated from previously described type strains of species within the genus Nonomuraea as shown in Table 1. Some differences between strain 10-20SHSuT and closely related type strains included the hydrolysis of starch, which was positive in strain 10-20SHSuT, whereas the other type strains showed negative result. Alkaline phosphatase production was negative in strain 10-20SHSuT, but other type strains were positive. Furthermore, the strain 10-20SHSuT was unable to produce oxidase, sensitive to lysozyme and grew at 43 °C contrary to N. soli JCM 17347T. Based on the above results, strain 10-20SHSuT should be classified as a representative of a novel species in the genus Nonomuraea, for which the name N. suaedae sp. nov. is proposed.
Description of Nonomuraea suaedae sp. nov
Nonomuraea suaedae (su.ae´dae. N. L. gen. fem. n. suaedae of the genus Suaeda, pertaining to the plant S. maritima (L.) Dumort).
Gram-stain-positive, aerobic and non-motile actinomycete that forms an extensively branched substrate and aerial mycelia. Spore chains are straight to flexuous. Spores are observed to be rod and non-motile with a smooth surfaces. Good growth is observed on ISP 2 and GYE agar, moderate growth on ISP 3, ISP 4, ISP 5 and ISP 7 but poor growth on ISP 6 agar. Colony colours vary from orange to deep orange. Brown soluble pigment is produced on ISP 2, ISP 3, ISP 4 and GYE agar but no soluble pigment is observed on ISP 5, ISP 6 and ISP 7. Grow at 15–43 °C (optimum 27–38 °C) and in the range of pH 5.0–9.0 (optimum pH 7.0–8.0). The maximum NaCl concentration for growth is 3% (w/v). Positive for production of catalase and nitrate reduction but negative for oxidase and H2S production. Sensitive to 0.005% lysozyme. Hydrolysis of aesculin, arbutin, carboxymethyl cellulose, casein, gelatin, hypoxanthine, starch and Tween 80 are positive, but negative for adenine, guanine, l-tyrosine, Tween 20, urea, xanthine and xylan. Utilizes l(+)arabinose, d(+)cellobiose, d(–)fructose, d(+)glucose, myo-inositol, d(–)lactose, maltose, d(+)mannose, d(–)mannitol, d(+)raffinose, l(+)rhamnose, d(–)ribose, sucrose, d(+)trehalose and d(+)xylose as sole carbon sources, but d(+)galactose, sodium citrate, sodium propionate, d(–)sorbitol and xylitol are not utilized. l-Asparagine, l-histidine and KNO3 are used as sole nitrogen sources. Shows activities of α-chymotrypsin, cystine arylamidase, esterase (C4), α-galactosidase, β-galactosidase, n-acetyl-β-glucosaminidase, α-glucosidase, β-glucosidase, leucine arylamidase, naphthol-AS-Bl-phosphohydrolase, trypsin and valine arylamidase. Acid phosphatase, alkaline phosphatase, esterase lipase (C8), α-fucosidase, β-glucoronidase, lipase (C14) and α-mannosidase are negative. Contains meso-diaminopimelic acid in cell-wall peptidoglycan. Galactose, glucose, madurose, mannose and ribose are present in whole-cell hydrolysates. MK-9(H4) is a major menaquinone of the organism. Polar lipids are diphosphatidylglycerol, hydroxy-phosphatidylethanolamine, hydroxy-phosphatidylmonomethylethanolamine, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol mannosides, phosphatidylinositol, phosphotidylmethylethanolamine, two unidentified sugar containing phosphoaminolipids and an unidentified phospholipid. The predominant cellular fatty acids are iso-C16:0, 10-methyl-C17:0 and C17:0.
The type strain is 10-20SHSuT (= TBRC 8487T = NBRC 113448T), which was isolated from the rhizosphere of S. maritima collected from Phetchaburi Province, Thailand. The DNA G + C content of the type strain is 71.9 mol%.
References
Zhang Z, Wang Y, Ruan J. Reclassification of Thermomonospora and Microtetraspora. Int J Syst Bacteriol. 1998;48:411–22.
Wu H, Liu B, Shao Y, Ou X, Huang F. Thermostaphylospora grisealba gen. nov., sp. nov., isolated from mushroom compost and transfer of Thermomonospora chromogena Zhang et al. 1998 to Thermostaphylospora chromogena comb. nov. Int J Syst Evol Microbiol. 2018;68:602–8.
Goodfellow M, Quintana E. Family 1. Streptosporangiaceae Goodfellow, Stanton, Simpson and Minnikin 1990a 321VP (Effective publication: Goodfellow, Stanton, Simpson and Minnikin 1990b.) emend. Ward-Rainey, Rainey and Stackebrandt 1997, 486 emend. Zhi, Li and Stackebrandt 2009, 600. In: Goodfellow M, et al. (eds). Bergey’s Manual® of Systematic Bacteriology. New York: Springer; 2012. p. 1807–11.
Nonomura H, Ohara Y. Distribution of actinomycetes in soil. XI. Some new species of the genus Actinomadura Lechevalier et al. J Ferment Technol. 1971;49:904–12.
Sungthong R, Nakaew N. The genus Nonomuraea: a review of a rare actinomycete taxon for novel metabolites. J Basic Microbiol. 2015;55:554–65.
Raju AS, Kumar R. On the reproductive ecology of Suaeda maritima, S. monoica and S. nudiflora (Chenopodiaceae). J Threat Taxa. 2016;8:8860–76.
Larsen K. Chenopodiaceae: Suaeda. In: Santisuk T, Larsen K, Nielsen I, Chayamarit K, Editors. Flora of Thailand. Bangkok, Thailand: The Forest Herbarium, Royal Forest Department; 2000. p. 257–9
Hayakawa M, Nonomura H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol. 1987;65:501–9.
Shirling ET, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–40.
Jacobson E, Grauville WC, Fogs CE. Color harmony manual. 4th ed. Chicago: Container Corporation of America; 1958.
Gordon RE, Barnett DA, Handerhan JE. Pang CH-N. Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol. 1974;24:54–63.
Williams S, et al. Numerical classification of Streptomyces and related genera. Microbiology. 1983;129:1743–813.
Küster E, Williams S. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–9.
Becker B, Lechevalier M, Lechevalier H. Chemical composition of cell-wall preparations from strains of various form-genera of aerobic actinomycetes. Appl Microbiol. 1965;13:236–43.
Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–22.
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–31.
Tomiyasu I. Mycolic acid composition and thermally adaptative changes in Nocardia asteroides. J Bacteriol. 1982;151:828–37.
Minnikin D, Patel P, Alshamaony L, Goodfellow M. Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Evol Microbiol. 1977;27:104–17.
Collins M, Pirouz T, Goodfellow M, Minnikin D. Distribution of menaquinones in actinomycetes and corynebacteria. Microbiology. 1977;100:221–30.
Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids. Newark, Delaware: MIDI, Inc; 2001. p. 1–7.
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical streptomyces genetics. Norwich, UK: John Innes Foundation; 2000.
Mingma R, Pathom-aree W, Trakulnaleamsai S, Thamchaipenet A, Duangmal K. Isolation of rhizospheric and roots endophytic actinomycetes from Leguminosae plant and their activities to inhibit soybean pathogen, Xanthomonas campestris pv. glycine. World J Microbiol Biotechnol. 2014;30:271–80.
Boratyn GM, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33.
Yoon S-H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–7.
Fitch WM. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst Biol. 1971;20:406–16.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Tamaoka J, Komagata K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett. 1984;25:125–8.
Ezaki T, Hashimoto Y, Yabuuchi E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Evol Microbiol. 1989;39:224–9.
Wayne L, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol. 1987;37:463–4.
Acknowledgements
This research was supported by Center of Excellence on Biodiversity (BDC), Office of Higher Education Commission (Project Code BDC-PG1-160003). We are grateful to Professor Dr Savitree Limtong of Kasetsart University, the director of research program. We acknowledge to The King’s Royally Initiated Leam Phak Bia Environmental Research and Development Project, Phetchaburi province, Thailand for samples collection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lipun, K., Teo, W.F.A., Tongpan, J. et al. Nonomuraea suaedae sp. nov., isolated from rhizosphere soil of Suaeda maritima (L.) Dumort. J Antibiot 72, 518–523 (2019). https://doi.org/10.1038/s41429-019-0159-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-019-0159-y
This article is cited by
-
Nonomuraea terrae sp. nov., isolated from arid soil
Archives of Microbiology (2020)