Abstract
Background
Differences in patients gut microbiota composition with the potential for dysbiosis have been associated with chronic kidney disease (CKD). However, factors other than the disease itself, such as diet and cohabitation, have not been evaluated when gut microbiota of CKD patients was compared with that of healthy controls. The aim of this study was to compare the gut microbiota composition between patients on peritoneal dialysis (PD) and age-matched household contacts with normal renal function.
Methods
Fecal samples were collected from 20 patients [men: 70%; age: 53.5 years (48.2–66; median and interquartile range); length on PD: 14 months (5.2–43.5) and 20 controls. The region V4 of the 16S ribosomal RNA gene was PCR-amplified and sequenced on Illumina MiSeq platform. Dietary intake and diet quality were assessed by a 3-day food record and a diet quality index, respectively.
Results
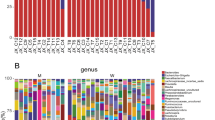
No difference was found between the gut microbiota composition of patients and controls, assessed by alpha and beta diversities (p > 0.05) and genera differential abundance (p > 0.05). The most abundant phyla in both groups were Firmicutes (PD = 45%; Control: 47%; p = 0.65) and Bacteroidetes (PD = 41%; Control: 45%; p = 0.17). The phylum Proteobacteria, known as a potential marker of gut dysbiosis, was not different in proportions between groups (p > 0.05). No difference was observed regarding diet quality and dietary intake of fiber, protein and other nutrients (p > 0.05).
Conclusion
Gut microbiota of patients on PD did not differ from household contacts. This result suggests that cohabitation and dietary intake might have outweighed the disease influence on gut microbiota composition of our PD patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Additional data are available from the corresponding author on reasonable request.
References
Relman DA. The human microbiome and the future practice of medicine. J Am Med Assoc. 2015;314:1127–8.
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8.
Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14.
Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7.
Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y, Keilbaugh SA, et al. Linking long-term dietary patterns with Gut Microbial Enterotypes. Science. 2011;334:105–8.
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Ley RE, Sogin ML, et al. A core gut microbiome between lean and obesity twins. Nature. 2009;457:480–4.
Dill-McFarland KA, Tang ZZ, Kemis JH, Kerby RL, Chen G, Palloni A, et al. Close social relationships correlate with human gut microbiota composition. Sci Rep. 2019;9:703.
van de Guchte M, Blottière HM, Doré J. Humans as holobionts: implications for prevention and therapy. Microbiome. 2018;6:81.
Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503.
Stadlbauer V, Horvath A, Ribitsch W, Schmerböck B, Schilcher G, Lemesch S, et al. Structural and functional differences in gut microbiome composition in patients undergoing haemodialysis or peritoneal dialysis. Sci Rep. 2017;7:15601.
Xu KY, Xia GH, Lu JQ, Chen MX, Zhen X, Wang S, et al. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci Rep. 2017;7:1445.
Hatch M, Vaziri ND. Enhanced enteric excretion of urate in rats with chronic renal failure. Clin Sci. 1994;86:511–6.
Kalantar-Zadeh K, Kopple JD, Deepak S, Block D, Block G. Food intake characteristics of hemodialysis patients as obtained by food frequency questionnaire. J Ren Nutr. 2002;12:17–31.
Cano AE, Neil AK, Kang JY, Barnabas A, Eastwood JB, Nelson SR, et al. Gastrointestinal symptoms in patients with end-stage renal disease undergoing treatment by hemodialysis or peritoneal dialysis. Am J Gastroenterol. 2007;102:1990–7.
Bammens B, Evenepoel P, Verbeke K, Vanrenterghem Y. Impairment of small intestinal protein assimilation in patients with end-stage renal disease: extending the malnutrition-inflammation-atherosclerosis concept. Am J Clin Nutr. 2004;80:1536–43.
Kortman GAM, Reijnders D, Swinkels DW. Oral iron supplementation: potential implications for the gut microbiome and metabolome in patients with CKD. Hemodial Int. 2017;21:S28–36.
Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long- term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836.
Jiang N, Zhang C, Feng H, Yuan J, Ding L, Fang W, et al. Clinical characteristics associated with the properties of gut microbiota in peritoneal dialysis patients. Perit Dial Int. 2021;41:298–306.
Caivano S, Colugnati FAB, Domene SMÁ. Diet quality index associated with digital food guide: update and validation. Cad Saude Publica. 2019;35:e00043419.
Kopple JD. National kidney foundation K/DOQI clinical practice guidelines for nutrition in chronic renal failure. Am J Kidney Dis. 2001;37:S66–70.
Drossman DA. Functional gastrointestinal disorders: history, pathophysiology, clinical features, and Rome IV. Gastroenterology. 2016;150:1262–79.
Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 1997;32:920–4.
Daugirdas JT. Second generation logarithmic estimates of single-pool variable volume Kt/V: an analysis of error. J Am Soc Nephrol. 1993;4:1205–13.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4.
Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Research. 2016;5:1492.
Renaud G, Stenzel U, Maricic T, Wiebe V, Kelso J. DeML: robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics. 2015;31:770–2.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol. 2017;261:169–76.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–40.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25.
Law CW, Chen Y, Shi W, Smyth GK. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Smyth GK Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3.
McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217.
Kim BR, Shin J, Guevarra RB, Lee JH, Kim DW, Seol KH, et al. Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol. 2017;27:2089–93.
Whittaker RH. Vegetation of the Siskiyou Mountains, Oregon and California. Ecol Monogr. 2013;30:279–338.
Claesson MJ, Jeffery IB, Conde S, Power SE, O’connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84.
Luo D, Zhao W, Lin Z, Wu J, Lin H, Li Y, et al. The effects of hemodialysis and peritoneal dialysis on the gut microbiota of end-stage renal disease patients, and the relationship between gut microbiota and patient prognoses. Front Cell Infect Microbiol. 2021;11:1–13.
Gao B, Alonzo-Palma N, Brooks B, Jose A, Barupal D, Jagadeesan M, et al. A pilot study on the effect of prebiotic on host-microbial co-metabolism in peritoneal dialysis patients. Kidney Int Reports. 2020;5:1309–15.
Hu X, Ouyang S, Xie Y, Gong Z, Du J. Characterizing the gut microbiota in patients with chronic kidney disease. Postgrad Med. 2020;132:495–505.
Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, Desantis TZ, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83:308–15.
Jiang S, Xie S, Lv D, Wang P, He H, Zhang T, et al. Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci Rep. 2017;7:1–10.
Wong J, Piceno YM, DeSantis TZ, Pahl M, Andersen GL, Vaziri ND. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am J Nephrol. 2014;39:230–7.
Nosratola DV, Ying-Yong Z, Madeleine VP. Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: The nature, mechanisms, consequences and potential treatment. Nephrol Dial Transplant. 2016;31:737–46.
Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2013:1–22.
Lax S, Smith DP, Hampton-marcell J, Owens SM, Handley M, Scott NM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–52.
Poesen R, Windey K, Neven E, Kuypers D, De Preter V, Augustijns P, et al. The influence of CKD on colonic microbial metabolism. J Am Soc Nephrol. 2016;27:1389–99.
García-Vega ÁS, Corrales-Agudelo V, Reyes A, Escobar JS. Diet quality, food groups and nutrients associated with the gut microbiota in a nonwestern population. Nutrients. 2020;12:1–21.
Tian XK, Wang T. A low-protein diet does not necessarily lead to malnutrition in peritoneal dialysis patients. J Ren Nutr. 2005;15:298–303.
Sutton D, Higgins B, Stevens JM. Continuous ambulatory peritoneal dialysis patients are unable to increase dietary intake to recommended levels. J Ren Nutr. 2007;17:329–35.
Acknowledgements
We gratefully acknowledge the nurses and all staff from Fundação Oswaldo Ramos (São Paulo, Brazil) for their assistance in data collection, the study participants and also to Danilo Takashi Aoike for his assistance in statistical analyses.
Funding
RRT, LSA and NBFP received a scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). LC receives a scholarship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (# 302765/2017-4). Support for this research was provided by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (# 2018/12122-7).
Author information
Authors and Affiliations
Contributions
The results presented in this paper have not been published previously in whole or part, except in abstract form. All authors contributed and approved the manuscript. RRT, LSA and LC conceived, designed, and implemented the study; RRT, LSA and NBFP collected the data; RRT performed the statistical analysis; RRT, LSA and LC interpreted the data, drafted the manuscript and were responsible for the final revisions. CH provided intellectual content of critical importance to the work and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The study protocol was approved by the Research Ethics Committee of the Universidade Federal de São Paulo (UNIFESP, São Paulo, Brazil, #1422/2018).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Teixeira, R.R., de Andrade, L.S., Pereira, N.B.F. et al. Gut microbiota profile of patients on peritoneal dialysis: comparison with household contacts. Eur J Clin Nutr 77, 90–97 (2023). https://doi.org/10.1038/s41430-022-01190-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41430-022-01190-7
This article is cited by
-
Correlation between gut microbiome and cognitive impairment in patients undergoing peritoneal dialysis
BMC Nephrology (2023)
-
Gut microbiota disturbances and protein-energy wasting in chronic kidney disease: a narrative review
Journal of Nephrology (2023)