Abstract
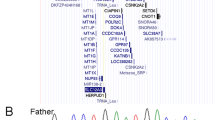
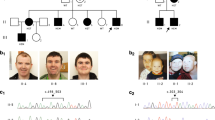
Here, we identified the causal mutation in the MRX20 family, one of the larger X-linked pedigrees that have been described in which no gene had been identified up till now. In 1995, the putative disease gene had been mapped to the pericentromeric region on the X chromosome, but no follow-up studies were performed. Here, whole exome sequencing (WES) on two affected and one unaffected family member revealed the c.195del/p.(Thr66ProfsTer55) mutation in the DLG3 gene (NM_021120.4) that segregated with the affected individuals in the family. DLG3 mutations have been consequently associated with intellectual disability and are a plausible explanation for the clinical abnormalities observed in this family. In addition, we identified two other variants co-segregating with the phenotype: a stop gain mutation in SSX1 (c.358G>T/p.(Glu120Ter)) (NM_001278691.2) and a nonsynonymous SNV in USP27X (c.56 A>G/p.(Gln19Arg)) (NM_001145073.3). RNA sequencing revealed 14 differentially expressed genes (p value < 0.1) in 7 affected males compared to 4 unaffected males of the family, including four genes known to be associated with neurological disorders. Thus, in this paper we identified the c.195del/p.(Thr66ProfsTer55) mutation in the DLG3 gene (NM_021120.4) as likely responsible for the phenotype observed in the MRX20 family.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated during the current study are not publicly available due consent restrictions, but are available from the corresponding author on reasonable request.
References
American Psychiatric Association, Diagnostic and statistical manual of mental disorders: DSM-5. 5th edition ed. Arlington, Va: American Psychiatric Association. xliv, 947 pages (2013).
Leonard H, Wen X. The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev. 2002;8:117–34.
Neri G, Schwartz CE, Lubs HA, Stevenson RE. X-linked intellectual disability update 2017. Am J Med Genet A. 2018;176:1375–88.
van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104.
Raymond FL. X linked mental retardation: a clinical guide. J Med Genet. 2006;43:193–200.
Lazzarini A, Stenroos ES, Lehner T, McKoy V, Gold B, McCormack MK, et al. Short tandem repeat polymorphism linkage studies in a new family with X-linked mental retardation (MRX20). Am J Med Genet. 1995;57:552–7.
Helsmoortel C, Vandeweyer G, Ordoukhanian P, Van Nieuwerburgh F, Van der Aa N, Kooy RF. Challenges and opportunities in the investigation of unexplained intellectual disability using family-based whole-exome sequencing. Clin Genet. 2015;88:140–8.
Vandeweyer G, Van Laer L, Loeys B, Van den Bulcke T, Kooy RF. VariantDB: a flexible annotation and filtering portal for next generation sequencing data. Genome Med. 2014;6:74.
Jones JR. Nonrandom X chromosome inactivation detection. Curr Protoc Hum Genet. 2014;80:9 7 1–9 7.
D’Incal CP, Cappuyns E, Choukri K, Szrama K, De Man K, Aa NVD, et al. In search of the hidden protein: optimization of detection strategies for autism-associated activity-dependent neuroprotective protein (ADNP) mutants. Res Square. 2022; https://doi.org/10.21203/rs.3.rs-1954095/v1.
Bushnell B. BBMap: a fast, accurate, splice-aware aligner. In Proceedings of 9th annual genomics of energy & environment meeting; United States (2014).
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205.
Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol. 2014;10:e1003731.
Liska O, Bohar B, Hidas A, Korcsmaros T, Papp B, Fazekas D, et al. TFLink: an integrated gateway to access transcription factor-target gene interactions for multiple species. Database. 2022;2022:baac083.
Iqbal Z, Vandeweyer G, van der Voet M, Waryah AM, Zahoor MY, Besseling JA, et al. Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet. 2013;22:1960–70.
Kumar R, Ha T, Pham D, Shaw M, Mangelsdorf M, Friend KL, et al. A non-coding variant in the 5′ UTR of DLG3 attenuates protein translation to cause non-syndromic intellectual disability. Eur J Hum Genet. 2016;24:1612–6.
Matis T, Michaud V, Van-Gils J, Raclet V, Plaisant C, Fergelot P, et al. Triple diagnosis of Wiedemann-Steiner, Waardenburg and DLG3-related intellectual disability association found by WES: a case report. J Gene Med. 2020;22:e3197.
Philips AK, Siren A, Avela K, Somer M, Peippo M, Ahvenainen M, et al. X-exome sequencing in Finnish families with intellectual disability-four novel mutations and two novel syndromic phenotypes. Orphanet J Rare Dis. 2014;9:49.
Tzschach A, Grasshoff U, Beck-Woedl S, Dufke C, Bauer C, Kehrer M, et al. Next-generation sequencing in X-linked intellectual disability. Eur J Hum Genet. 2015;23:1513–8.
Tarpey P, Parnau J, Blow M, Woffendin H, Bignell G, Cox C, et al. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:318–24.
Zanni G, van Esch H, Bensalem A, Saillour Y, Poirier K, Castelnau L, et al. A novel mutation in the DLG3 gene encoding the synapse-associated protein 102 (SAP102) causes non-syndromic mental retardation. Neurogenetics. 2010;11:251–5.
Sandestig A, Green A, Aronsson J, Ellnebo K, Stefanova M. A novel DLG3 mutation expanding the phenotype of X-linked intellectual disability caused by DLG3 nonsense variants. Mol Syndromol. 2020;10:281–5.
Gieldon L, Mackenroth L, Betcheva-Krajcir E, Rump A, Beck-Wodl S, Schallner J, et al. Skewed X-inactivation in a family with DLG3-associated X-linked intellectual disability. Am J Med Genet A. 2017;173:2545–50.
Plenge RM, Stevenson RA, Lubs HA, Schwartz CE, Willard HF. Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. Am J Hum Genet. 2002;71:168–73.
Muller BM, Kistner U, Kindler S, Chung WJ, Kuhlendahl S, Fenster SD, et al. SAP102, a novel postsynaptic protein that interacts with NMDA receptor complexes in vivo. Neuron. 1996;17:255–65.
Consortium GT. Erratum: Genetic effects on gene expression across human tissues. Nature. 2018;553:530.
Lopez-Hernandez A, Sberna S, Campaner S. Emerging principles in the transcriptional control by YAP and TAZ. Cancers. 2021;13:4242.
Hayashi R, Goto Y, Ikeda R, Yokoyama KK, Yoshida K. CDCA4 is an E2F transcription factor family-induced nuclear factor that regulates E2F-dependent transcriptional activation and cell proliferation. J Biol Chem. 2006;281:35633–48.
Liu H, Ippolito GC, Wall JK, Niu T, Probst L, Lee BS, et al. Functional studies of BCL11A: characterization of the conserved BCL11A-XL splice variant and its interaction with BCL6 in nuclear paraspeckles of germinal center B cells. Mol Cancer. 2006;5:18.
Beckmann H, Su LK, Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990;4:167–79.
Dias C, Estruch SB, Graham SA, McRae J, Sawiak SJ, Hurst JA, et al. BCL11A haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am J Hum Genet. 2016;99:253–74.
Lehalle D, Vabres P, Sorlin A, Bierhals T, Avila M, Carmignac V, et al. De novo mutations in the X-linked TFE3 gene cause intellectual disability with pigmentary mosaicism and storage disorder-like features. J Med Genet. 2020;57:808–19.
Paulraj P, Bosworth M, Longhurst M, Hornbuckle C, Gotway G, Lamb AN, et al. A novel homozygous deletion within the FRY gene associated with nonsyndromic developmental delay. Cytogenet Genome Res. 2019;159:19–25.
Huang Q, Zhang YF, Li LJ, Dammer EB, Hu YB, Xie XY, et al. Adult-onset neuronal ceroid lipofuscinosis with a novel DNAJC5 mutation exhibits aberrant protein palmitoylation. Front Aging Neurosci. 2022;14:829573.
Sans N, Petralia RS, Wang YX, Blahos J 2nd, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–71.
Won S, Levy JM, Nicoll RA, Roche KW. MAGUKs: multifaceted synaptic organizers. Curr Opin Neurobiol. 2017;43:94–101.
Cuthbert PC, Stanford LE, Coba MP, Ainge JA, Fink AE, Opazo P, et al. Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J Neurosci. 2007;27:2673–82.
Chen BS, Gray JA, Sanz-Clemente A, Wei Z, Thomas EV, Nicoll RA, et al. SAP102 mediates synaptic clearance of NMDA receptors. Cell Rep. 2012;2:1120–8.
Wei Z, Wu G, Chen BS. Regulation of SAP102 synaptic targeting by phosphorylation. Mol Neurobiol. 2018;55:6215–26.
Roszkowska M, Krysiak A, Majchrowicz L, Nader K, Beroun A, Michaluk P, et al. SRF depletion in early life contributes to social interaction deficits in the adulthood. Cell Mol Life Sci. 2022;79:278.
Chaiwongkot A, Kitkumthorn N, Srisuttee R, Buranapraditkun S. Cellular expression profiles of Epstein-Barr virus-transformed B-lymphoblastoid cell lines. Biomed Rep. 2020;13:43.
Liu C, Si W, Tu C, Tian S, He X, Wang S, et al. Deficiency of primate-specific SSX1 induced asthenoteratozoospermia in infertile men and cynomolgus monkey and tree shrew models. Am J Hum Genet. 2023;110:516–30.
McBride MJ, Mashtalir N, Winter EB, Dao HT, Filipovski M, D’Avino AR, et al. The nucleosome acidic patch and H2A ubiquitination underlie mSWI/SNF recruitment in synovial sarcoma. Nat Struct Mol Biol. 2020;27:836–45.
Centore RC, Sandoval GJ, Soares LMM, Kadoch C, Chan HM. Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet. 2020;36:936–50.
Hu H, Haas SA, Chelly J, Van Esch H, Raynaud M, de Brouwer AP, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry. 2016;21:133–48.
Kobayashi T, Iwamoto Y, Takashima K, Isomura A, Kosodo Y, Kawakami K, et al. Deubiquitinating enzymes regulate Hes1 stability and neuronal differentiation. FEBS J. 2015;282:2411–23.
Acknowledgements
We thank Cheryl S. Reid for family consultation.
Funding
The authors acknowledge the support of the Research Fund of the University of Antwerp OEC-Methusalem grant “GENOMED”.
Author information
Authors and Affiliations
Contributions
Clinical examination and counseling of the family were performed by AL and MKM. JH was responsible for the conceptualization and overview of the experiments under the supervision of RFK and GV. Primers were developed by JH and EE. Experiments were executed by JH, EE, KEVR, and BC. GV analyzed the WES data. Differential expression was analyzed by LM. Pathway enrichment analysis and identification of enriched transcription factors were performed by JH and LM. Analysis of RT-PCR data and preparation of the corresponding figures were performed by JH and CPD. Western Blotting was performed by CPD. JH drafted the manuscript, which was reviewed and approved by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The cell lines used in this study were donated with informed consent to the Human Genetic Mutant Cell Repository at the Coriell Institute (Camden, New Jersey, USA) for research purposes.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huyghebaert, J., Mateiu, L., Elinck, E. et al. Identification of a DLG3 stop mutation in the MRX20 family. Eur J Hum Genet 32, 317–323 (2024). https://doi.org/10.1038/s41431-024-01537-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-024-01537-7
This article is cited by
-
Solving medical mysteries with genomics
European Journal of Human Genetics (2024)