Abstract
The mechanisms that restrict regeneration and maintain cell identity following injury are poorly characterized in higher vertebrates. Following β-cell loss, 1–2% of the glucagon-producing α-cells spontaneously engage in insulin production in mice. Here we explore the mechanisms inhibiting α-cell plasticity. We show that adaptive α-cell identity changes are constrained by intra-islet insulin- and Smoothened-mediated signalling, among others. The combination of β-cell loss or insulin-signalling inhibition, with Smoothened inactivation in α- or δ-cells, stimulates insulin production in more α-cells. These findings suggest that the removal of constitutive ‘brake signals’ is crucial to neutralize the refractoriness to adaptive cell-fate changes. It appears that the maintenance of cell identity is an active process mediated by repressive signals, which are released by neighbouring cells and curb an intrinsic trend of differentiated cells to change.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The RNA-Seq data generated in this study have been deposited in the NCBI GEO database under accession number GSE109285. Source data for Figs. 1–6 and Supplementary Figs. 1,2,4–6 are provided as Supplementary Table 1 and RNA analyses are provided in Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Holmberg, J. & Perlmann, T. Maintaining differentiated cellular identity. Nat. Rev. Genet. 13, 429–439 (2012).
Thowfeequ, S., Myatt, E. J. & Tosh, D. Transdifferentiation in developmental biology, disease, and in therapy. Dev. Dynam. 236, 3208–3217 (2007).
Chera, S. & Herrera, P. L. Regeneration of pancreatic insulin-producing cells by in situ adaptive cell conversion. Curr. Opin. Genet. Dev. 40, 1–10 (2016).
Pasque, V., Jullien, J., Miyamoto, K., Halley-Stott, R. P. & Gurdon, J. B. Epigenetic factors influencing resistance to nuclear reprogramming. Trends Genet. 27, 516–525 (2011).
Natoli, G. Maintaining cell identity through global control of genomic organization. Immunity 33, 12–24 (2010).
Barrero, M. J., Boue, S. & Izpisua Belmonte, J. C. Epigenetic mechanisms that regulate cell identity. Cell Stem Cell 7, 565–570 (2010).
Szabat, M. et al. Maintenance of beta-cell maturity and plasticity in the adult pancreas: developmental biology concepts in adult physiology. Diabetes 61, 1365–1371 (2012).
Eade, K. T. & Allan, D. W. Neuronal phenotype in the mature nervous system is maintained by persistent retrograde bone morphogenetic protein signaling. J. Neurosci. 29, 3852–3864 (2009).
Lopez-Coviella, I., Berse, B., Krauss, R., Thies, R. S. & Blusztajn, J. K. Induction and maintenance of the neuronal cholinergic phenotype in the central nervous system by BMP-9. Science 289, 313–316 (2000).
Jessen, K. R., Mirsky, R. & Arthur-Farraj, P. The role of cell plasticity in tissue repair: adaptive cellular reprogramming. Dev. Cell 34, 613–620 (2015).
Knapp, D. & Tanaka, E. M. Regeneration and reprogramming. Curr. Opin. Genet. Dev. 22, 485–493 (2012).
Chera, S. et al. Diabetes recovery by age-dependent conversion of pancreatic delta-cells into insulin producers. Nature 514, 503–507 (2014).
Thorel, F. et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464, 1149–1154 (2010).
Chakravarthy, H. et al. Converting adult pancreatic islet alpha cells into beta Cells by targeting both Dnmt1 and Arx. Cell. Metab. 25, 622–634 (2017).
Bosma, M. J. & Carroll, A. M. The SCID mouse mutant: definition, characterization, and potential uses. Annu. Rev. Immunol. 9, 323–350 (1991).
Yang, Y. P., Thorel, F., Boyer, D. F., Herrera, P. L. & Wright, C. V. Context-specific alpha- to-beta-cell reprogramming by forced Pdx1 expression. Genes Dev. 25, 1680–1685 (2011).
Schaffer, L. et al. A novel high-affinity peptide antagonist to the insulin receptor. Biochem. Biophys. Res. Commun. 376, 380–383 (2008).
Vikram, A. & Jena, G. S961, an insulin receptor antagonist causes hyperinsulinemia, insulin-resistance and depletion of energy stores in rats. Biochem. Biophys. Res. Commun. 398, 260–265 (2010).
Yi, P., Park, J. S. & Melton, D. A. Betatrophin: a hormone that controls pancreatic beta cell proliferation. Cell 153, 747–758 (2013).
Cigliola, V., Thorel, F., Chera, S. & Herrera, P. L. Stress-induced adaptive islet cell identity changes. Diabetes Obes. Metab. 18, 87–96 (2016).
Damond, N. et al. Blockade of glucagon signaling prevents or reverses diabetes onset only if residual beta-cells persist. eLife 5, e13828 (2016).
Grieco, F. A. et al. Delta-cell-specific expression of hedgehog pathway Ptch1 receptor in murine and human endocrine pancreas. Diabetes Metab. Res. Rev. 27, 755–760 (2011).
Lau, J. & Hebrok, M. Hedgehog signaling in pancreas epithelium regulates embryonic organ formation and adult beta-cell function. Diabetes 59, 1211–1221 (2010).
Kawahira, H. et al. Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development 130, 4871–4879 (2003).
Cervantes, S., Lau, J., Cano, D. A., Borromeo-Austin, C. & Hebrok, M. Primary cilia regulate Gli/Hedgehog activation in pancreas. Proc. Natl Acad. Sci. USA 107, 10109–10114 (2010).
Landsman, L., Parent, A. & Hebrok, M. Elevated Hedgehog/Gli signaling causes beta-cell dedifferentiation in mice. Proc. Natl Acad. Sci. USA 108, 17010–17015 (2011).
Thomas, M. K., Lee, J. H., Rastalsky, N. & Habener, J. F. Hedgehog signaling regulation of homeodomain protein islet duodenum homeobox-1 expression in pancreatic beta-cells. Endocrinology 142, 1033–1040 (2001).
Thomas, M. K., Rastalsky, N., Lee, J. H. & Habener, J. F. Hedgehog signaling regulation of insulin production by pancreatic beta-cells. Diabetes 49, 2039–2047 (2000).
Schaffer, A. E. et al. Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. PLoS Genet. 9, e1003274 (2013).
Gao, T. et al. Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metab. 19, 259–271 (2014).
Gutierrez, G. D. et al. Pancreatic beta cell identity requires continual repression of non-beta cell programs. J. Clin. Invest. 127, 244–259 (2017).
Swisa, A. et al. PAX6 maintains beta cell identity by repressing genes of alternative islet cell types. J. Clin. Invest. 127, 230–243 (2017).
Papizan, J. B. et al. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev. 25, 2291–2305 (2011).
Gauthier, B. R., Gosmain, Y., Mamin, A. & Philippe, J. The beta-cell specific transcription factor Nkx6.1 inhibits glucagon gene transcription by interfering with Pax6. Biochem. J. 403, 593–601 (2007).
Schisler, J. C. et al. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet beta cells. Proc. Natl Acad. Sci. USA 102, 7297–7302 (2005).
Lee, Y. et al. Hyperglycemia in rodent models of type 2 diabetes requires insulin-resistant alpha cells. Proc. Natl Acad. Sci. USA 111, 13217–13222 (2014).
Butler, A. E. et al. Adaptive changes in pancreatic beta cell fractional area and beta cell turnover in human pregnancy. Diabetologia 53, 2167–2176 (2010).
White, M. G. et al. Expression of mesenchymal and alpha-cell phenotypic markers in islet beta-cells in recently diagnosed diabetes. Diabetes Care 36, 3818–3820 (2013).
Thorel, F. et al. Normal glucagon signaling and beta-cell function after near-total alpha-cell ablation in adult mice. Diabetes 60, 2872–2882 (2011).
Hayashi, Y. et al. Mice deficient for glucagon gene-derived peptides display normoglycemia and hyperplasia of islet α-cells but not of intestinal L-cells. Mol. Endocrinol. 23, 1990–1999 (2009).
Miyatsuka, T. et al. Persistent expression of PDX-1 in the pancreas causes acinar-to-ductal metaplasia through Stat3 activation. Genes Dev. 20, 1435–1440 (2006).
Dietrich, P., Dragatsis, I., Xuan, S., Zeitlin, S. & Efstratiadis, A. Conditional mutagenesis in mice with heat shock promoter-driven cre transgenes. Mamm. Genome 11, 196–205 (2000).
Bruning, J. C. et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 2, 559–569 (1998).
Long, F., Zhang, X. M., Karp, S., Yang, Y. & McMahon, A. P. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128, 5099–5108 (2001).
Chen, S. X. et al. Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis. Model Mech. 4, 537–547 (2011).
Rodriguez, C. I. et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25, 139–140 (2000).
Russell, M. C., Cowan, R. G., Harman, R. M., Walker, A. L. & Quirk, S. M. The hedgehog signaling pathway in the mouse ovary. Biol. Reprod. 77, 226–236 (2007).
Li, Z. et al. Reduced white fat mass in adult mice bearing a truncated Patched 1. Int. J. Biol. Sci. 4, 29–36 (2008).
Strom, A. et al. Unique mechanisms of growth regulation and tumor suppression upon Apc inactivation in the pancreas. Development 134, 2719–2725 (2007).
Mathe, Z. et al. Tetracycline-regulated expression of VEGF-A in beta cells induces angiogenesis: improvement of engraftment following transplantation. Cell Transplant. 15, 621–636 (2006).
Acknowledgements
We are grateful to Y. Dor, A. Ruiz i Altaba and M. González Gaitán for carefully reading and discussing the manuscript; to O. Fazio, M. Urwyler, C. Gysler, B. Polat and R. Gangula for their technical help and to J.-P. Aubry-Lachainaye for FACS assistance. We acknowledge C. Delucinge-Vivier, M. Docquier, A. Efanov, P. Ebert, J. Calley, H. Wu, S.K. Syed and T. Wei for technical and expert assistance supporting RNA sequencing, quality control, statistical and bioinformatics analyses. M.S. was supported by R01DK068471 (NIH/NIDDK). S.C. is supported by grants from the Research Council of Norway (NFR no. 247577) and the Novo Nordisk Foundation (no. NNF15OC0015054). M.A.M. was supported by U01DK072473 (Beta Cell Biology Consortium). Work was funded by grants from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Disease (Beta Cell Biology Consortium, grant no. U01DK089566 and Human Islet Research Network, grant nos UC4-DK104209 and UC4-DK108132), the Juvenile Diabetes Research Foundation (in part through joint research funding with Lilly Research Laboratories, grant nos 17-2011-276, and 2-SRA-2015-67-Q-R), the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 155005 (IMIDIA), resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution, the Fondation privée des Hôpitaux Universitaires de Genève–Confirm, the Fondation Aclon and the Swiss National Science Foundation (grant no. NRP63, 310030_152965 and the Bonus of Excellence grant no.310030B_173319) to P.L.H.
Author information
Authors and Affiliations
Contributions
V.C., F.T., L.G., S.C., D.B. and K.F. performed all of the experiments and most analyses. T.M., H.K, C.E.V.W. and M.S. provided transgenic lines. M.K.T., S.G., S.C., K.F. and L.V.G analysed RNA-Seq data. M.A.M. and A.B.O. generated the Sst-rtTA knock-in mouse line. K.F., F.T., V.C., L.G., S.C., D.O. and P.L.H. conceived the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
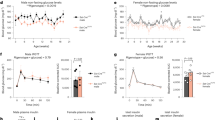
Supplementary Figure 1 Insulin production by α-cells after β-cell ablation in islets transplanted under the kidney capsule.
(a) Immunofluorescence staining of insulin (green) and glucagon (red) in RIP-DTR islets transplanted under the kidney capsule, either before (‘no DT’) or after (‘DT’) β-cell ablation. The scatter graph reports a quantification of the β-cell ablation efficiency, measured as the number of insulin-containing cells remaining per islet section in non-treated and DT-treated animals, 10 days later. (mean +/− s.e.m., n = 40 islet sections per condition. (b) Experimental design of Exp. #4, where islets were transplanted in the kidney capsule of immunocompromised mice (SCID) either mixed together or at 2 separated sites. (c) Proportion of YFP-traced α-cells expressing insulin 1-month post-DT in mixed RIP-DTR+WT islets compared to RIP-DTR islets from 2-sides graft. Data are shown as mean +/− s.d.; n = 3 biologically independent animals per condition. (d) Immunofluorescence staining of mCherry (magenta), YFP (green), insulin (red) in transplanted mixed RIP-DTR+WT islets 1-month post-DT. Scale bars: 20 µm. Experiment repeated independently 3 times. See Supplementary Table 1 as source data.
Supplementary Figure 2 FACS sorting. Gating strategy used for FACS sorting the different islet cell types used for RNA seq analyses.
Mice used to purify the different islet cell types are listed in the methods.
Supplementary Figure 3 α-cell transcriptome analysis and gene expression changes after β-cell loss.
(a) PCA plot of all samples analyzed by RNA-Seq. Native β-cells (‘β’, n = 5 mice) show a different profile compared to α-cell groups including native α-cells (‘α’, n = 6 mice), α-cells 1 month after DT-induced β-cell ablation (‘αDT’, n = 3 mice), α-cells overexpressing Pdx1 (‘αPdx1OE’, n = 3 mice), and α-cells overexpressing Pdx1 combined with β-cell ablation (‘αPdx1OE+DT’, n = 5 mice). (b) Heat map showing scaled expression (blue, high; white, low) of α- / β-cell-enriched genes differentially expressed in αDT, αPdx1OE, αPdx1OE+DT conditions compared to native α-cells. (c) Gene expression changes (log2 value) in αPdx1OE+DT (top), αPdx1OE (middle) and αDT (bottom) compared to native α-cells. DEGs indicated by red-colored background show induced β-cell signature upon the conditions, while DEGs indicated by green-colored background show refractory responses. * FDR < 0.05. n = 6 mice in α, n = 5 mice in αPdx1OE+DT, n = 3 mice in αPdx1OE, and n = 3 mice in αDT. (d) Transgenes required for constitutive labeling α-cells with the reporter Venus and experimental design for α-cell labeling and purification after β-cell loss. (e) RT-qPCR analyses for different components of the insulin/IGF1-R signaling pathway. Components of the pathway are downregulated in α-cells 5 days post-DT. Center indicates the mean. qPCR were performed in triplicates and data processing and statistical analyses were performed using the web-based software ‘RT2 Profiler PCR Array Data Analysis version 3.5’, from Qiagen. Quantification of the gene expression changes were performed using the 2−ΔΔCt method. P-values were calculated using a Student’s t-test (two-tail distribution and equal variances between the two samples) on the 2−ΔCt values. Genes with P<0.05 are shown. n = 3 mice. qPCR was performed once. (f) Experimental design for DT-mediated β-cell loss, islet isolation and qPCR. Igf1 mRNA is upregulated in islets after β-cell loss, likely as an attempt to sustain insulin signaling. Data shown as mean +/− sem; n = 3 mice with 3 technical replicates each. Center indicates the mean. Two-tailed unpaired t-test P = 0.0086. (g) Gene set enrichment analysis (GSEA) from RNA-Seq showing enrichment of the insulin signaling pathway components PKC (left) and PI3K (right) in α-cells after DT-mediated β-cell ablation (αDT) compared to native α-cells (α). P-values were calculated by an empirical phenotype-based permutation test. n = 6 mice in α, n = 3 mice in αDT. NES: normalized enrichment score. See Supplementary Table 2 as source data.
Supplementary Figure 4 Insulin production in α-cells under insulin signaling blockade.
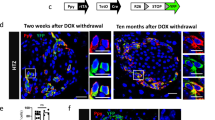
(a) Transgenes for inducible α-cell lineage tracing and experimental design for α-cell lineage tracing and S961 treatment. (b-c) Immunofluorescence staining for the β-cell-specific transcription factors Pdx1 and Nkx6.1 in islets of mice treated for 1 month with S961. Experiments were repeated once, with n = 3 mice treated asynchronously according to their availability. (d) Mice and experimental design for α-cell labeling and sorting by FACS. (e) β-cell-specific genes are upregulated in α-cells upon S961 treatment. qPCRs were performed in triplicates. Data shown as mean +/− sem; n = 3 mice. Experiment performed once. Two-tailed unpaired t-test (P = 0.0017 INS1; P = 0.0010 INS2) (f) Transgenes required for inducible α-cell lineage tracing, simultaneous insulin receptor (IR) and IGF-1 receptor (IGF1R) downregulation, and ectopic Pdx1 induction in adult α-cells. (g) Impaired insulin/IGF1 signaling in α-cells through IR and IGF1R downregulation predisposes α-cells to insulin gene expression in mice with normal β-cell mass. (h) Fractions of α-cells expressing insulin after IR/IGF1R downregulation in α-cells and Pdx1OE. n = 3 mice. Two-tailed Mann Whitney test, P = 0.05. (i) Transgenes required for inducible Pdx1 expression in adult α-cells. (j) Wortmannin-mediated PI3K inhibition or S961-mediated insulin antagonism trigger insulin production in Pdx1OE α-cells. Insulin production in Pdx1OE α-cells is reversed when islets are analyzed 1 month after ending S961 treatment. (k) Fraction of converted α-cells found upon insulin signaling antagonism/inhibition. Black and red symbols: control and αPdx1OE mice, respectively. n = 6, 4, 3, 3 mice in DOX only, DOX+Wort, DOX+S961 and DOX+S961+STOP, respectively. Two-tailed Mann Whitney test, P = 0.0061 Pdx1 only vs Pdx1+wort, P = 0.0167 Pdx1 only vs Pdx1+S961, P = 0.0333 Pdx1 only vs Pdx1+S961then stop. All experiments were performed once with animals treated in an asynchronous way depending on their availability. Scale bars: 20 μm. See Supplementary Table 1d as source data.
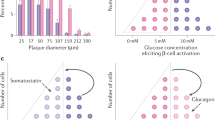
Supplementary Figure 5 Hedgehog signaling modulation in α- and δ-cells.
(a) Hh signaling components are enriched in α- vs β-cells and their expression is unchanged 30 days upon DT-induced β-cell ablation, Pdx1OE or both. Log2FC are taken from the RNA-Seq presented in Fig. 3. (b) Indian Hedgehog ligand expression in islets of control mice. Immunofluorescence was repeated two times with similar results. (c) Non-quantitative RT-PCR of Hedgehog pathway’s components in sorted α-, δ- and β-cells. Liver, duodenum and uterus were used as controls. Experiments were repeated three times with similar results. Unprocessed scans of the blots are provided in Supplementary Fig. 7. (d) Transgenes required for simultaneous α-cell lineage tracing, Smo co-receptor downregulation and DT-induced β-cell ablation. (e) Experimental design. (f) Smo and (g) α-cell genes Arx and Glucagon are downregulated in α-cells with reduced Smo activity upon DOX treatment. n = 3 mice. qPCR was performed once. Two-tailed unpaired t-test; SMO: P = 0.0012, GCG: P = 0.0325 fl/+ vs +/+ P = 0.0141 fl/fl vs +/+ and P = 0.0177 fl/fl vs fl/+, ARX: P = 0.0214 fl/+ vs +/+, 0.0245 fl/fl vs +/+). Center indicates the mean. (h) Smo inactivation in α-cells in absence of β-cell loss or insulin signaling impairment does not lead to insulin production in intact islets. Immunofluorescence was repeated once, on 2 slides of 3 different mice. (i) Transgenes required to lineage trace δ-cells, experimental design and characterization of labeling specificity in the Sst-rtTA line. Only δ-cells express YFP. n = 5, 4, 4 for δ-, β- and α-cells, respectively. (j) Transgenes required for lineage tracing and Smo coreceptor inactivation in δ-cells, and for DT-induced β-cell ablation and experimental design. Smo is downregulated in purified δ-cells after DOX treatment. n = 3 mice. qPCRs were performed once. Two-tailed unpaired t-test, P = 0.0048. Downregulation of Smo in δ-cells does not alter the expression of the δ-cell genes Somatostatin and Hhex. n = 3 mice. qPCR was performed once. Center indicates the mean. Insulin protein is not expressed in δ-cells upon Smo inactivation in intact islets. Immunofluorescence was repeated once, on 3 slides of 3 different mice. (k) Experimental design for δ-cell lineage tracing and DT-induced β-cell loss in δ-Smo-KO mice. A fraction of δ-cells express insulin after β-cell ablation in δ-Smo-KO mice, but their percentage is not increased when compared to mice with intact Smo expression. n = 3, 3, 3 mice in no DT wt/wt, wt/fl, fl/fl respectively and n = 4, 4, 3 mice in DT wt/wt, wt/fl, fl/fl respectively. Immunofluorescence was repeated once, on 3 to 5 slides per mouse. Center in the graph indicates the mean. Two-tailed unpaired t-test. (l) Transgenes required for α-cell tracing and DT-induced β-cell ablation, experimental design and immunofluorescence staining of mouse islets 1 month after DT+GANT61 treatment. Immunofluorescence was repeated once, on 3 to 5 slides of n = 8 mice treated with DT+GANT61. A representative islet is shown. (m) The % of α-cells expressing insulin (n = 5 and 4 mice for DT+DMSO and DT+GANT61, respectively), the number of insulin-expressing cells per islet section (n = 8 mice for DT+DMSO and DT+GANT61), and the % of islets containing insulin+ cells (n = 8 mice and n = 9 mice for DT+DMSO and DT+GANT61, respectively) are increased in mice treated with GANT61 after DT-induced β-cell loss. Two-tailed unpaired t-test (P = 0.0236 panel left, P = 0.0085 middle, P = 0.0213 right graph. (n) Transgenes required for simultaneous β- and δ-cell ablation, experimental design and percentage of cells coexpressing insulin and glucagon after β- and δ-cell loss. n = 4, 5, 3 mice for β-, β+δ and δ-cell ablation, respectively. Center indicates the mean. Two tailed unpaired t-test, P = 0.0494. Scale bars: 10 μm. See Supplementary Table 1m,n,q as source data.
Supplementary Figure 6 Constitutive signals ensure maintenance of cell identity.
In physiological conditions, the identity of fully differentiated cells (such as α-cells) is maintained through a variety of constitutive signals preventing cell identity changes. Most of these brakes (f.i. insulin signaling) are preserved upon stress and injury (f.i. β-cell loss). However, few cells may undergo changes in cell identity as an attempt to replenish the compromised cell population (f.i. α-cells produce insulin to cope with insulin insufficiency). Due to the persistence of most constitutive brakes, this kind of natural cell interconversion events are rare and help explaining the poor regenerative capacity observed in higher vertebrates. Insulin-expressing α-cells may reacquire their initial fate upon restoration of physiological conditions (reversibility). Future therapies aimed at identifying and abrogating constitutive regeneration brakes should improve the intrinsic regenerative capacity of likely any organ.
Supplementary Figure 7
Unprocessed scans of the non-quantitative PCR blots provided in Supplementary Fig. 6.
Supplementary information
Supplementary Information
Supplementary Figures 1–7 and Supplementary Table legends.
Supplementary Table 1
Statistics source data
Supplementary Table 2
RNA analyses
Supplementary Table 3
Primers.
Supplementary Table 4
Antibodies.
Rights and permissions
About this article
Cite this article
Cigliola, V., Ghila, L., Thorel, F. et al. Pancreatic islet-autonomous insulin and smoothened-mediated signalling modulate identity changes of glucagon+ α-cells. Nat Cell Biol 20, 1267–1277 (2018). https://doi.org/10.1038/s41556-018-0216-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0216-y
This article is cited by
-
Keeping pace: the primary cilium as the conducting baton of the islet
Diabetologia (2024)
-
Novel time-resolved reporter mouse reveals spatial and transcriptional heterogeneity during alpha cell differentiation
Diabetologia (2024)
-
Islet cell replacement and transplantation immunology in a mouse strain with inducible diabetes
Scientific Reports (2022)
-
STAT3 suppression and β-cell ablation enhance α-to-β reprogramming mediated by Pdx1
Scientific Reports (2022)
-
Stage-specific transcriptomic changes in pancreatic α-cells after massive β-cell loss
BMC Genomics (2021)