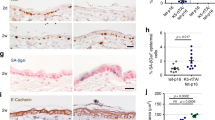
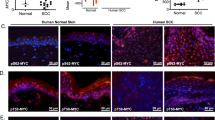
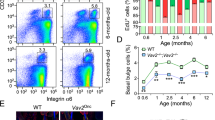
Abstract
Oncogenic lesions are surprisingly common in morphologically and functionally normal human skin. However, the cellular and molecular mechanisms that suppress their cancer-driving potential to maintain tissue homeostasis are unknown. By employing assays for the direct and quantitative assessment of cell fate choices in vivo, we show that oncogenic activation of PI3K–AKT, the most commonly activated oncogenic pathway in cancer, promotes the differentiation and cell cycle exit of epidermal progenitors. As a result, oncogenic PI3K–AKT-activated epidermis exhibits a growth disadvantage even though its cells are more proliferative. We then sought to uncover the underlying mechanism behind oncogene-induced differentiation via a series of genetic screens in vivo. An AKT substrate, SH3RF1, is identified as a specific promoter of epidermal differentiation that has no effect on proliferation. Our study provides evidence for a direct, cell autonomous mechanism that can suppresses progenitor cell renewal and block clonal expansion of epidermal cells bearing a common and activating mutation in Pik3ca.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA–seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE99659. Previously published microarray data that were re-analysed here are available under accession codes GSE4170434, GSE4885935, GSE2605936 and GSE5053070. Source data for Figs. 1e, 2b–d, f, 3d, g, 4a, c, e, 5g, h, j, 6c, e, 7g, i and Supplementary Figs. 2c, 3a, d, 4a–c, e, 5c–e, i, 7a–g, i, j are provided in Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Pardal, R., Molofsky, A. V., He, S. & Morrison, S. J. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb. Symp. Quant. Biol. 70, 177–185 (2005).
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Michaloglou, C. et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 (2005).
Braig, M. et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005).
Chen, Z. et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 (2005).
Fearnhead, H. O. et al. Oncogene-dependent apoptosis is mediated by caspase-9. Proc. Natl Acad. Sci. USA 95, 13664–13669 (1998).
Lowe, S. W., Cepero, E. & Evan, G. Intrinsic tumour suppression. Nature 432, 307–315 (2004).
Bissonnette, R. P., Echeverri, F., Mahboubi, A. & Green, D. R. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature 359, 552–554 (1992).
Fuchs, E. & Raghavan, S. Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 3, 199–209 (2002).
Jones, K. B. & Klein, O. D. Oral epithelial stem cells in tissue maintenance and disease: the first steps in a long journey. Int. J. Oral Sci. 5, 121–129 (2013).
Tomasetti, C. & Levy, D. Role of symmetric and asymmetric division of stem cells in developing drug resistance. Proc. Natl Acad. Sci. USA 107, 16766–16771 (2010).
Manning, B. D. & Cantley, L. C. AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 (2007).
Zumsteg, Z. S. et al. Taselisib (GDC-0032), a potent beta-sparing small molecule inhibitor of PI3K, radiosensitizes head and neck squamous carcinomas containing activating PIK3CA alterations. Clin. Cancer Res. 22, 2009–2019 (2016).
Mizrachi, A. et al. Tumour-specific PI3K inhibition via nanoparticle-targeted delivery in head and neck squamous cell carcinoma. Nat. Commun. 8, 14292 (2017).
Martins, F., de Sousa, S. C., Dos Santos, E., Woo, S. B. & Gallottini, M. PI3K-AKT-mTOR pathway proteins are differently expressed in oral carcinogenesis. J. Oral Pathol. Med. 45, 746–752 (2016).
Watanabe, S. et al. Activation of PI3K-AKT pathway in oral epithelial dysplasia and early cancer of tongue. Bull. Tokyo Dent. Coll. 50, 125–133 (2009).
Liu, S. et al. The PI3K-Akt pathway inhibits senescence and promotes self-renewal of human skin-derived precursors in vitro. Aging Cell 10, 661–674 (2011).
Pankow, S., Bamberger, C., Klippel, A. & Werner, S. Regulation of epidermal homeostasis and repair by phosphoinositide 3-kinase. J. Cell Sci. 119, 4033–4046 (2006).
Calautti, E., Li, J., Saoncella, S., Brissette, J. L. & Goetinck, P. F. Phosphoinositide 3-kinase signaling to Akt promotes keratinocyte differentiation versus death. J. Biol. Chem. 280, 32856–32865 (2005).
Janes, S. M. et al. PI3-kinase-dependent activation of apoptotic machinery occurs on commitment of epidermal keratinocytes to terminal differentiation. Cell Res. 19, 328–339 (2009).
Gunschmann, C. et al. Insulin/IGF-1 controls epidermal morphogenesis via regulation of FoxO-mediated p63 inhibition. Dev. Cell 26, 176–187 (2013).
Dainichi, T. et al. PDK1 is a regulator of epidermal differentiation that activates and organizes asymmetric cell division. Cell Rep. 15, 1615–1623 (2016).
Segrelles, C. et al. Akt signaling leads to stem cell activation and promotes tumor development in epidermis. Stem Cells 32, 1917–1928 (2014).
Murayama, K. et al. Akt activation induces epidermal hyperplasia and proliferation of epidermal progenitors. Oncogene 26, 4882–4888 (2007).
Beronja, S., Livshits, G., Williams, S. & Fuchs, E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 16, 821–827 (2010).
Beronja, S. et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 501, 185–190 (2013).
Vanhaesebroeck, B., Stephens, L. & Hawkins, P. PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 13, 195–203 (2012).
Li, Y. Y. et al. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin. Cancer Res. 21, 1447–1456 (2015).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Kinross, K. M. et al. An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. J. Clin. Invest. 122, 553–557 (2012).
Tikoo, A. et al. Physiological levels of Pik3ca(H1047R) mutation in the mouse mammary gland results in ductal hyperplasia and formation of ERalpha-positive tumors. PLoS ONE 7, e36924 (2012).
Hare, L. M. et al. Physiological expression of the PI3K-activating mutation Pik3ca(H1047R) combines with Apc loss to promote development of invasive intestinal adenocarcinomas in mice. Biochem. J. 458, 251–258 (2014).
Blanpain, C., Lowry, W. E., Geoghegan, A., Polak, L. & Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 (2004).
De Craene, B. et al. Epidermal Snail expression drives skin cancer initiation and progression through enhanced cytoprotection, epidermal stem/progenitor cell expansion and enhanced metastatic potential. Cell Death Differ. 21, 310–320 (2014).
Radoja, N., Gazel, A., Banno, T., Yano, S. & Blumenberg, M. Transcriptional profiling of epidermal differentiation. Physiol. Genomics 27, 65–78 (2006).
Rendl, M., Polak, L. & Fuchs, E. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 22, 543–557 (2008).
Adam, R. C. et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370 (2015).
Williams, S. E., Beronja, S., Pasolli, H. A. & Fuchs, E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470, 353–358 (2011).
Fuchs, E. Epidermal differentiation and keratin gene expression. J. Cell. Sci. Suppl. 17, 197–208 (1993).
Rompolas, P. et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474 (2016).
Hornbeck, P. V. et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 (2012).
Figueroa, C., Tarras, S., Taylor, J. & Vojtek, A. B. Akt2 negatively regulates assembly of the POSH-MLK-JNK signaling complex. J. Biol. Chem. 278, 47922–47927 (2003).
de Bock, C. E. et al. Protein interaction screening identifies SH3RF1 as a new regulator of FAT1 protein levels. FEBS Lett. 591, 667–678 (2017).
Tuvia, S. et al. The ubiquitin E3 ligase POSH regulates calcium homeostasis through spatial control of Herp. J. Cell Biol. 177, 51–61 (2007).
Xu, Z., Kukekov, N. V. & Greene, L. A. POSH acts as a scaffold for a multiprotein complex that mediates JNK activation in apoptosis. EMBO J. 22, 252–261 (2003).
Xu, Z., Sproul, A., Wang, W., Kukekov, N. & Greene, L. A. Siah1 interacts with the scaffold protein POSH to promote JNK activation and apoptosis. J. Biol. Chem. 281, 303–312 (2006).
Lyons, T. R. et al. Regulation of the pro-apoptotic scaffolding protein POSH by Akt. J. Biol. Chem. 282, 21987–21997 (2007).
Xu, Z. & Greene, L. A. Activation of the apoptotic JNK pathway through the Rac1-binding scaffold protein POSH. Methods Enzymol. 406, 479–489 (2006).
Weston, C. R. et al. The c-Jun NH2-terminal kinase is essential for epidermal growth factor expression during epidermal morphogenesis. Proc. Natl Acad. Sci. USA 101, 14114–14119 (2004).
Zhang, J. Y., Green, C. L., Tao, S. & Khavari, P. A. NF-kappaB RelA opposes epidermal proliferation driven by TNFR1 and JNK. Genes Dev. 18, 17–22 (2004).
Xu, Z., Maroney, A. C., Dobrzanski, P., Kukekov, N. V. & Greene, L. A. The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Mol. Cell. Biol. 21, 4713–4724 (2001).
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012).
Marshall, C. J. Tumor suppressor genes. Cell 64, 313–326 (1991).
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Munoz-Espin, D. & Serrano, M. Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 15, 482–496 (2014).
Podsypanina, K. et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl Acad. Sci. USA 96, 1563–1568 (1999).
Li, G. et al. Conditional loss of PTEN leads to precocious development and neoplasia in the mammary gland. Development 129, 4159–4170 (2002).
Du, L. et al. Overexpression of PIK3CA in murine head and neck epithelium drives tumor invasion and metastasis through PDK1 and enhanced TGFβ signaling. Oncogene 35, 4641–4652 (2016).
Fuchs, E. Scratching the surface of skin development. Nature 445, 834–842 (2007).
Watt, F. M., Lo Celso, C. & Silva-Vargas, V. Epidermal stem cells: an update. Curr. Opin. Genet. Dev. 16, 518–524 (2006).
van der Flier, L. G. & Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241–260 (2009).
Hao, J. J. et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 48, 1500–1507 (2016).
Beronja, S. & Fuchs, E. RNAi-mediated gene function analysis in skin. Methods Mol. Biol. 961, 351–361 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 15, 550 (2014).
Schramek, D. et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343, 309–313 (2014).
Yu, X. & Zuo, Y. Two-photon in vivo imaging of dendritic spines in the mouse cortex using a thinned-skull preparation.J. Vis. Exp. 87, e51520 (2014).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Gozdecka, M. et al. JNK suppresses tumor formation via a gene-expression program mediated by ATF2. Cell Rep. 9, 1361–1374 (2014).
Acknowledgements
The authors thank the following: W. Phillips for sharing the inducible Pik3caH1047R mouse model; V. Greco and C. Pineda for sharing the photoactivatable mouse model and the intravital imaging technique; M. Schober and A. Hsieh for critical reading of the manuscript; the Comparative Medicine Department (AAALAC-accredited; R. Uthamanthil, Director) for care of the mice in accordance with National Institutes of Health (NIH) guidelines; the Genomics Department (J. Delrow, Director) for sequencing; the Scientific Imaging Department (J. Vazquez, Director) for advice; and the Flow Cytometry Department (A. Berger, Director) for flow cytometry and FACS. This research was supported by grants from the NIH (R01-AR070780 to S.B.), a Cell & Molecular Biology Training Grant (to M.S.), and a Thomsen Family Fellowship (to Z.Y.). The authors dedicate this work to X. Chen, and are grateful for her strength and support.
Author information
Authors and Affiliations
Contributions
Z.Y. and S.B. conceived the study and designed the experiments. Z.Y. performed all the experiments and data analyses. M.S. developed the EdU–BrdU differentiation assay. Z.Y. and M.S. performed the intravital mouse imaging. Z.Y. and S.B. wrote the paper. All authors provided intellectual input, vetted and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Gene targeting approach to model common oncogenic lesions in vivo.
(a), Catalog of 33 most common cancer-driver genes with oncogenic lesions (copy number alteration and mutation) across squamous cell carcinomas (SCC). (b), Lentiviral constructs for expression of ORFs and shRNAs. Barcode in the pLX EF1 vector and shRNA in the pLKO1 vector are used for sequencing-based identification and quantification of individual gene-targeting constructs. (c), Correlation between protein expression of the V5-tagged ORF library (V5/Actin) in 293 T cells and primary mouse keratinocytes (KRT). Control ORF (GFP) is in blue. (d), Ten most commonly mutated oncogenes in epithelial cancer. Statistics are based on TCGA data for carcinomas of the bladder, bowel, breast, head & neck, kidney, lung, ovary, pancreas, cervix, esophagus, stomach, liver, prostate, thymus, thyroid and uterus tissues. SCCs included SCC of the head & neck, lung and cervix. Each cancer type was equally weighted in the statistics.
Supplementary Figure 2 Oncogenic Pik3ca promotes cell division.
(a), Epidermal transduction with LV-Cre results in efficient activation of Pik3caH1047R expression. (b), Western blot demonstrates increased phosphorylation of AKT in LV-Cre transduced skin epithelium expressing PIK3CAH1047. Experiment was repeated 3 times independently with similar results. Unprocessed blots see Supplementary Fig. 8. (c), Epidermal expression of PIK3CAH1047R results in faster cell division at all stages of tissue growth and homeostasis. Statistics based on n = 3 animals of each condition. Error bar: SD, center value: mean. (d), Schematic comparison of how theoretical differences in the length of individual cell cycle stages may affect EdU-BrdU pulse chase labeling, and how to calculate cell division interval from the observed numbers. Assay specifically labels cells which complete S-phase during the initial 2hrs of EdU pulse. The length of the G1/S phase length does not, while significant arrest in G2 may affect the number of EdU+ only cells at the assay time point. Statistical source data for (c) are shown in Supplementary Table 4.
Supplementary Figure 3 EdU-BrdU pulse chase differentiation assay measures renewal rate in epidermis.
(a) and (b), Statistics (a) and representative images (b) show that short (2 hour) pulse of EdU results in cell labeling that is restricted to the K10- progenitor cell population in wild type (WT) and oncogenic Pik3ca-expressing epidermis. Statistics based on n = 3 animals of each condition. Error bar: SD, center value: mean. Scale bar, 50 μm. (c) and (d), Representative flow plot (c) and statistics (d) show that in the EdU+ only population isolated from EdU-BrdU pulse-chase labeled epidermis, only ~3% cells are arrested at G2 (2n population), and there is no significant difference in the size of the G2 arrested population in WT and Pik3ca 2X epidermis. Statistics based on n = 3 WT animals and 5 Pik3ca 2X animals. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. (e), Representative cell labeling in the EdU-BrdU pulse-chase assay. Experiment was repeated 3 times independently with similar results. EdU marks cells that have replicated their DNA over the 8 hours of the experiment. BrdU labels replicating cells during the last 6 hours of it. EdU+ only expression marks daughter cells that were given sufficient time to commit to progenitor or differentiated fate, as reported by expression of differentiation marker K10. Scale bar, 50 μm. Statistical source data for (a,d) are shown in Supplementary Table 4.
Supplementary Figure 4 Oncogenic Pik3ca regulates progenitor cell renewal and proliferation.
(a-c), Clonal and ubiquitous (field) activated Pik3caH1047R expression in skin does not affect epithelial cell crowding (a), and results in equal rates of renewal (b) and cell division (c). However, the rate of renewal is elevated in WT cells adjacent to Pik3ca 2X clone ( ≤ 5 cells from Pik3ca 2X clone) but not in WT cells that are non-adjacent to Pik3ca 2X clones. Statistics based on n = 3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. (d), Staining of differentiation marker K10, loricrin and filaggrin exhibited a similar pattern in WT and Pik3ca 2X tissue. Experiment was repeated 3 times independently with similar results. Scale bar, 50 μm. e, Long term EdU chase assay started at P21 shows that tissue turnover rates are similar between WT and Pik3ca 2X epidermis. Statistics based on n = 3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. (f), GSEA shows that oncogenic Pik3ca biases gene expression in epidermal progenitors towards differentiation. Statistics are based on n = 3 animals of each condition. ES, NES, FDR generated by GSEA2 program. Statistical source data for (a-c,e) are shown in Supplementary Table 4.
Supplementary Figure 5 Oncogenic Pik3ca promote differentiation via suppression of progenitor self-renewal.
(a), Set-up for live imaging of head skin epidermis. (b), Visualization of LV-Cre-mGFP transduced cells in vivo. Arrows indicate hair follicles. Suprabasal cells are identified based on their flattened morphology. Basal cells are identified based on their cuboidal morphology and proximity to the basement membrane (blue; second harmonic generation signal from Collagen). Scale bar, 50 μm. Experiment was repeated in transduced head skin of 4 animals independently with similar results. (c-e), Activation rate and bias of LV-CreER transduced epidermis treated with low dosage of Tamoxifen. At P20, CreER activity, based on conversion of mT to mG, is detected in ~1% of epidermal cells (c), majority of clones (~90%) are found as single cells (d), and most of the single cells ( > 98%) are localized to the basal/progenitor layer (e). Statistics based on n = 7 WT animals and 6 Pik3ca 2X animals. Error bar: SD, center value: mean. (f), FACS plot of GFP+/- cells sorted from Pik3ca 2X R26mT/mG epidermis transduced with LV-Cre or with LV-CreER and treated with low dose of Tamoxifen. Experiment was repeated 3 times independently with similar results. (g), Schematic of genotyping primers31 detecting substitution of wild type exon 20 with H1047R exon 20 of Pik3ca. (h), Genotyping PCR shows that low dose of Tamoxifen can efficiently activate Pik3caH1047R. Experiment was repeated 3 times independently with similar results. (i), Rate of progenitor cell renewal in WT and Pik3ca 2X epidermis observed using direct two-photon imaging is consistent with EdU-BrdU pulse-chase assay quantification. Statistics based on n = 3 animals of each condition. One-way ANOVA, P-value as indicated. Error bar: SD, center value: mean.
Supplementary Figure 6 Suppressed progenitor renewal rate predicts long-term loss of oncogenic Pik3ca-expressing clones.
(a), Schematic of clonal growth in epidermis started from a single cell with rate of renewal = 0.5, division interval = 1 day, and tissue turnover rate = 10 days. (b), Theoretical growth pattern of epidermal clones started from a single basal cell, and with a constant division interval and tissue turnover rate = 10 days (based on long term EdU chase and previous publication41). Rate of progenitor renewal is in the physiological range of 0.4-0.6. (c), Theoretical growth pattern of wild type and Pik3ca 2X clones started as a single cell at P20. Rates of differentiation, proliferation and tissue turnover are as observed in 3 independent differentiation and tissue turnover assays.
Supplementary Figure 7 Oncogenic PI3K/AKT signaling regulates JNK signaling through SH3RF1.
(a), mRNA expression of Akt1-3 in keratinocytes transduced with control shScram or gene-targeting shRNAs. Two shRNAs for each gene that induced strongest transcript depletion (marked with red arrow) were used for functional studies in vivo. Experiment was repeated 3 times independently with similar results. Error bar: SD, center value: mean. (b-e), Depletion of Akt1-3 shows that Akt1 and Akt2 are required for epidermal differentiation and cell division phenotypes. EdU-BrdU pulse-chase assay shows that Akt1 and Akt2 are required for differentiation in Pik3ca 2 × (b) and WT (c) skin epithelium. Cell division rate is equally dependent on Akt1 and Akt2 in Pik3ca 2 × (d) and WT (e) epidermis. Statistics based on n = 3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. (f), Sh3rf1 mRNA expression in keratinocytes transduced with gene-targeting shRNAs relative to control shScram. Two most efficient shRNAs (red arrows) were used in subsequent gene-depletion studies. Experiment was repeated 3 times independently with similar results. Error bar: SD, center value: mean. (g), SH3RF1 mutants do not affect cell division rate in both wild type and Pik3ca 2X epidermis. Statistics based on n = 3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean. (h), Immunofluorescence staining of phosphorylated JNK (red) in clonally activated Pik3ca 2X epidermis (green). Transduction with LV-Cre reduces p-JNK levels (top rows) that are rescued by overexpression of Sh3rf1 (bottom rows). Inset boundaries are marked by white rectangle in larger view images. Dotted line marks epidermal/dermal boundary. Dashed line marks the interface between Cre-transduced cells. Scale bar, 25 μm. Experiment was repeated 3 times independently with similar results. (i), MLK1 does not affect cell division rate in Pik3ca 2X epidermis. (j), Knockdown of Jnk1 does not affect cell division rate in Pik3ca 2X epidermis expressing SH3RF1SA. Statistics based on n = 3 animals of each condition. Two-tailed t test, P-value as indicated. Error bar: SD, center value: mean.
Supplementary Figure 8
Uncropped Western blots presented in the current study.
Supplementary information
Supplementary Information
Supplementary Figures 1–8 and legends for Supplementary Tables 1–4
Rights and permissions
About this article
Cite this article
Ying, Z., Sandoval, M. & Beronja, S. Oncogenic activation of PI3K induces progenitor cell differentiation to suppress epidermal growth. Nat Cell Biol 20, 1256–1266 (2018). https://doi.org/10.1038/s41556-018-0218-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0218-9
This article is cited by
-
Injury prevents Ras mutant cell expansion in mosaic skin
Nature (2023)
-
Metabolic determinants of tumour initiation
Nature Reviews Endocrinology (2023)
-
pTINCR microprotein promotes epithelial differentiation and suppresses tumor growth through CDC42 SUMOylation and activation
Nature Communications (2022)
-
The polarity protein PARD3 and cancer
Oncogene (2021)
-
Extracellular serine controls epidermal stem cell fate and tumour initiation
Nature Cell Biology (2020)