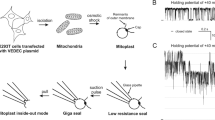
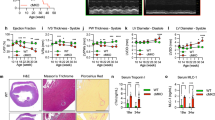
Abstract
Mitochondria provide chemical energy for endoergonic reactions in the form of ATP, and their activity must meet cellular energy requirements, but the mechanisms that link organelle performance to ATP levels are poorly understood. Here we confirm the existence of a protein complex localized in mitochondria that mediates ATP-dependent potassium currents (that is, mitoKATP). We show that—similar to their plasma membrane counterparts—mitoKATP channels are composed of pore-forming and ATP-binding subunits, which we term MITOK and MITOSUR, respectively. In vitro reconstitution of MITOK together with MITOSUR recapitulates the main properties of mitoKATP. Overexpression of MITOK triggers marked organelle swelling, whereas the genetic ablation of this subunit causes instability in the mitochondrial membrane potential, widening of the intracristal space and decreased oxidative phosphorylation. In a mouse model, the loss of MITOK suppresses the cardioprotection that is elicited by pharmacological preconditioning induced by diazoxide. Our results indicate that mitoKATP channels respond to the cellular energetic status by regulating organelle volume and function, and thereby have a key role in mitochondrial physiology and potential effects on several pathological processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ashcroft, F. M., Harrison, D. E. & Ashcroft, S. J. Glucose induces closure of single potassium channels in isolated rat pancreatic β-cells. Nature 312, 446–448 (1984).
Ashcroft, F. M. & Rorsman, P. KATP channels and islet hormone secretion: new insights and controversies. Nat. Rev. Endocrinol. 9, 660–669 (2013).
Nichols, C. G. KATP channels as molecular sensors of cellular metabolism. Nature 440, 470–476 (2006).
Inoue, I., Nagase, H., Kishi, K. & Higuti, T. ATP-sensitive K+ channel in the mitochondrial inner membrane. Nature 352, 244–247 (1991).
Paucek, P. et al. Reconstitution and partial purification of the glibenclamide-sensitive, ATP-dependent K+ channel from rat liver and beef heart mitochondria. J. Biol. Chem. 267, 26062–26069 (1992).
Garlid, K. D. & Halestrap, A. P. The mitochondrial KATP channel—fact or fiction? J. Mol. Cell. Cardiol. 52, 578–583 (2012).
Augustynek, B. B., Kunz, W. S. & Szewczyk, A. in Pharmacology of Mitochondria (Handbook of Experimental Pharmacology vol. 240) (eds Singh, H. & Sheu, S.S.) 103–127 (Springer, Cham, 2016).
Nessa, A., Rahman, S. A. & Hussain, K. Hyperinsulinemic hypoglycemia – the molecular mechanisms. Front. Endocrinol. (Lausanne) 7, 29 (2016).
Garlid, K. D. et al. Cardioprotective effect of diazoxide and its interaction with mitochondrial ATP-sensitive K+ channels. Possible mechanism of cardioprotection. Circ. Res. 81, 1072–1082 (1997).
O’Rourke, B. Evidence for mitochondrial K+ channels and their role in cardioprotection. Circ. Res. 94, 420–432 (2004).
Sato, T., Sasaki, N., Seharaseyon, J., O’Rourke, B. & Marbán, E. Selective pharmacological agents implicate mitochondrial but not sarcolemmal KATP channels in ischemic cardioprotection. Circulation 101, 2418–2423 (2000).
Wojtovich, A. P. et al. Kir6.2 is not the mitochondrial KATP channel but is required for cardioprotection by ischemic preconditioning. Am. J. Physiol. Heart Circ. Physiol. 304, H1439–H1445 (2013).
Szabo, I. & Zoratti, M. Mitochondrial channels: ion fluxes and more. Physiol. Rev. 94, 519–608 (2014).
Smith, C. O., Nehrke, K. & Brookes, P. S. The Slo(w) path to identifying the mitochondrial channels responsible for ischemic protection. Biochem. J. 474, 2067–2094 (2017).
Calvo, S. E., Clauser, K. R. & Mootha, V. K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 (2016).
The GTEx Consortium. The Genotype–Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015).
Dahlem, Y. A. et al. The human mitochondrial KATP channel is modulated by calcium and nitric oxide: a patch-clamp approach. Biochim. Biophys. Acta 1656, 46–56 (2004).
Bednarczyk, P. et al. Quinine inhibits mitochondrial ATP-regulated potassium channel from bovine heart. J. Membr. Biol. 199, 63–72 (2004).
Choma, K. et al. Single channel studies of the ATP-regulated potassium channel in brain mitochondria. J. Bioenerg. Biomembr. 41, 323–334 (2009).
Cang, C., Aranda, K., Seo, Y. J., Gasnier, B. & Ren, D. TMEM175 is an organelle K+ channel regulating lysosomal function. Cell 162, 1101–1112 (2015).
Schaedler, T. A. et al. Structures and functions of mitochondrial ABC transporters. Biochem. Soc. Trans. 43, 943–951 (2015).
Ardehali, H., Chen, Z., Ko, Y., Mejía-Alvarez, R. & Marbán, E. Multiprotein complex containing succinate dehydrogenase confers mitochondrial ATP-sensitive K+ channel activity. Proc. Natl Acad. Sci. USA 101, 11880–11885 (2004).
Lee, S.-Y. et al. Architecture mapping of the inner mitochondrial membrane proteome by chemical tools in live cells. J. Am. Chem. Soc. 139, 3651–3662 (2017).
Bernardi, P. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol. Rev. 79, 1127–1155 (1999).
Liu, X. & Hajnóczky, G. Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death Differ. 18, 1561–1572 (2011).
Duchen, M. R., Leyssens, A. & Crompton, M. Transient mitochondrial depolarizations reflect focal sarcoplasmic reticular calcium release in single rat cardiomyocytes. J. Cell Biol. 142, 975–988 (1998).
Schwarzländer, M. et al. Pulsing of membrane potential in individual mitochondria: a stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis. Plant Cell 24, 1188–1201 (2012).
Wang, W. et al. Superoxide flashes in single mitochondria. Cell 134, 279–290 (2008).
Rosselin, M., Santo-Domingo, J., Bermont, F., Giacomello, M. & Demaurex, N. L-OPA1 regulates mitoflash biogenesis independently from membrane fusion. EMBO Rep. 18, 451–463 (2017).
Leanza, L. et al. Direct pharmacological targeting of a mitochondrial ion channel selectively kills tumor cells in vivo. Cancer Cell 31, 516–531.e10 (2017).
Garlid, K. D. & Paucek, P. The mitochondrial potassium cycle. IUBMB Life 52, 153–158 (2001).
Patten, D. A. et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 33, 2676–2691 (2014).
Costa, A. D. T. & Garlid, K. D. Intramitochondrial signaling: interactions among mitoKATP, PKCε, ROS, and MPT. Am. J. Physiol. Heart Circ. Physiol. 295, H874–H882 (2008).
Varanita, T. et al. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab. 21, 834–844 (2015).
Liu, Y., Sato, T., O’Rourke, B. & Marban, E. Mitochondrial ATP-dependent potassium channels: novel effectors of cardioprotection? Circulation 97, 2463–2469 (1998).
Walters, A. M., Porter, G. A. Jr & Brookes, P. S. Mitochondria as a drug target in ischemic heart disease and cardiomyopathy. Circ. Res. 111, 1222–1236 (2012).
Foster, D. B. et al. Mitochondrial ROMK channel is a molecular component of mitoKATP. Circ. Res. 111, 446–454 (2012).
Ye, J. et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134 (2012).
Carraretto, L. et al. A thylakoid-located two-pore K+ channel controls photosynthetic light utilization in plants. Science 342, 114–118 (2013).
Teardo, E. et al. Physiological characterization of a plant mitochondrial calcium uniporter in vitro and in vivo. Plant Physiol. 173, 1355–1370 (2017).
Ashok, Y., Nanekar, R. & Jaakola, V.-P. Defining thermostability of membrane proteins by western blotting. Protein Eng. Des. Sel. 28, 539–542 (2015).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Frezza, C., Cipolat, S. & Scorrano, L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protocols 2, 287–295 (2007).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Granatiero, V., Patron, M., Tosatto, A., Merli, G. & Rizzuto, R. Using targeted variants of aequorin to measure Ca2+ levels in intracellular organelles. Cold Spring Harb. Protoc. 2014, 86–93 (2014).
Nicholls, D. G. et al. Bioenergetic profile experiment using C2C12 myoblast cells. J. Vis. Exp. 3–7, 2511 (2010).
Carpi, A. et al. The cardioprotective effects elicited by p66Shc ablation demonstrate the crucial role of mitochondrial ROS formation in ischemia/reperfusion injury. Biochim. Biophys. Acta 1787, 774–780 (2009).
Di Lisa, F., Menabò, R., Canton, M., Barile, M. & Bernardi, P. Opening of the mitochondrial permeability transition pore causes depletion of mitochondrial and cytosolic NAD+ and is a causative event in the death of myocytes in postischemic reperfusion of the heart. J. Biol. Chem. 276, 2571–2575 (2001).
Schlüter, K. D., Schwartz, P., Siegmund, B. & Piper, H. M. Prevention of the oxygen paradox in hypoxic-reoxygenated hearts. Am. J. Physiol. 261, H416–H423 (1991).
Acknowledgements
The authors are grateful to P. Bernardi, V. Petronilli, T. Pozzan, L. Scorrano and M. Zoratti for helpful discussion, to F. Caicci and F. Boldrin for electron microscopy, to A. Montagna for immunofluorescence, to L. Carraretto for help with expression of MITOK in E. coli, to L. Cendron for the help with thermal shift assay, and to M. Ruzzene and L. Cesaro for the help with the radioactive assay. This work was supported by grants from the University of Padova (Assegno junior 2015 and SID 2016 to D.D.S., STARS@UNIPD WiC grant 2017 to R.R. and UNIPD funds for research equipment 2015), the Italian Ministry of Education, University and Research (FIRB to R.R. PRIN no. 2015795S5W to I.S.), the European Union (ERC mitoCalcium, no. 294777 to R.R.), NIH (Grant no. 1P01AG025532-01A1 to R.R.), the Italian Association for Cancer Research (AIRC IG18633 to R.R. and IG20286 to I.S.), and Telethon-Italy (GGP16029 to R.R.).
Author information
Authors and Affiliations
Contributions
F.D.L. designed and discussed ischaemia–reperfusion experiments. R.M. and G.D.M. performed and analysed ischaemia–reperfusion experiments. I.S. designed the electrophysiological study. V.C. performed recordings, and I.S. and V.C. analysed biophysical data. R.R. and D.D.S designed and supervised all other experiments. A.P., A.C. and D.D.S. performed all other experiments and analysed data. I.S., R.R. and D.D.S conceived the study, discussed all the results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
: The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Diana Stojanovski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Overexpression of mouse MITOK causes mitochondrial dysfunction.
a, Membrane (pellet) and soluble (supernatant) proteins were separated from isolated liver mitochondria using ice-cold 0.1 M Na2CO3 (pH 11.5). Western blot is representative of three independent experiments. b, Proteinase K protection assay in isolated liver mitochondria. Similar results were obtained in three independent reactions. c, Mitochondrial morphology of control and MITOK-overexpressing HeLa cells. Representative of five independent experiments. Scale bar, 10 μm. d, Representative images and average ± s.d. traces of control and MITOK–GFP-expressing HeLa cells loaded with TMRM. n = 9 biologically replicates from 3 independent experiments. e, [Ca2+]mt measurements (mean ± s.d.) in intact HeLa cells that express the indicated constructs. n = 4 biological replicates, representative of 3 independent experiments. *P < 0.001 using one-way ANOVA with Holm–Sidak correction. f, qPCR analyses of transcripts from HeLa cells or the indicated human tissues using specific (isoform 1 and isoform 2) or non-specific (pan) primer pairs for MITOK. Data are normalized to ACTB and expressed as mean ± s.d. For HeLa cells, n = 3 biologically independent samples. For human tissues, n = 3 technical replicates. g, Protein expression of MITOK isoforms in HeLa cells transfected with the indicated constructs. Asterisk indicates a non-specific band. Image is representative of two independent experiments.
Extended Data Fig. 2 Localization and function of human MITOK isoforms.
a, Immunolocalization of MITOK (green) and the mitochondrial marker HSP60 (cyan) in HeLa cells transfected with the indicated constructs. Images are representative of two independent experiments. Scale bar, 10 μm. b, [Ca2+]mt measurements (mean ± s.d.) in intact HeLa cells that express the indicated constructs. n ≥ 6 independent samples, *P < 0.001 using one-way ANOVA with Holm–Sidak correction.
Extended Data Fig. 3 MITOK is a cation channel.
a, b, Western blots and Coomassie staining of MITOK expressed and purified from E. coli (a) or WGL (b). Representative of three independent experiments. c, Current traces showing two channels gating together, resulting in flickering activity (top), or normal single-channel activity (bottom). The two traces were recorded in the same experiment, performed in 100 mM K-gluconate medium. Representative of five independent experiments. d, Current trace showing burst-like activity. Recording was performed in 100 mM K-gluconate medium. Similar activity was present in more than six recordings. e, Representative traces of MITOK channel activity at the indicated voltages. Similar results were obtained in three independent experiments. f, Voltage ramp (from -120 mV to 0 mV) of MITOK recorded in 100 mM K-gluconate symmetrical medium (n = 3 independent experiments). g, Single-channel I–V curves under symmetric (black) and asymmetric (grey) ionic conditions. Mean value ± s.d., n = 50 from 3–4 different experiments for each point. Fitting revealed an Erev = –21.6 ± 1 mV. n = 4 independent experiments. h, Representative traces (top) and amplitude histograms (bottom) before and after the addition of 2 mM Ba2+ (n = 4 independent experiments) in 100 mM KCl medium. i, Representative traces (top) and amplitude histograms (bottom) of MITOK activity before (control) and after addition of paxilline (40 μM). n = 4 independent experiments.
Extended Data Fig. 4 MITOK alone is insensitive to ATP.
a, Activity of MITOK in 100 mM Na-gluconate. n = 5 independent experiments. b, Current traces of MITOK channel activity obtained from 60-s recordings (in 100 mM K-gluconate) before (top) and after (bottom) addition of 2 mM Mg and ATP. Representative of eight independent experiments. Voltages of the cis side are reported. c, Representative traces (left) and amplitude histograms (right) of MITOK activity before (control) and after the addition of 5-HD (100 μM). n = 5 independent experiments.
Extended Data Fig. 5 Biophysical characterization of recombinant human MITOSUR and mouse MITOK.
a–c, Thermal shift assay analysis of human MITOSUR and mouse MITOK. Average curves (a), graphs of Taggr (b, expressed as mean ± s.d.) and western blot (c). Representative of four independent experiments. d, Membrane extraction and western blot (representative of two independent experiments) of in vitro co-expressed human MITOSUR and mouse MITOK incorporated into liposomes. e, f, Membrane topology assessed by proteinase K protection assay in reconstituted liposomes and probed for mouse MITOK (e) and human MITOSUR (f). g, The same experiment shown in Fig. 2a is here represented with a different time scale. h, Amplitude histograms of channel activity before (control) and after first addition of 500 μM Mg and ATP, the second addition of 500 μM Mg and ATP and the third addition of 80 μM diazoxide. Similar results were obtained with four independent preparations. Open probabilities over a period of 120 s were: control, 0.62; 0.5 mM Mg/ATP, 0.14; 1 mM Mg/ATP, 0; diazoxide, 0.75. i, Single-channel current (I)–voltage (V) relationship of human MITOSUR and mouse MITOK. Linear fitting revealed a chord conductance of 63 ± 3 pS. n = 4 independent experiments. j, Activity in the absence (control, top) and presence of 1 mM Mg2+ (bottom) in 100 mM K-gluconate medium. n = 3 independent experiments. k, Activity of human MITOSUR and mouse MITOK in 100 mM K-gluconate, 5 mM EDTA, 10 mM HEPES, pH 7.4 n = 3 independent experiments. l, Human MITOSUR and mouse MITOK channel activity in 100 mM Na-gluconate medium. n = 4 independent experiments.
Extended Data Fig. 6 MITOK and MITOSUR interact in situ.
a, Proteinase K protection assay in isolated HeLa mitochondria. Similar results were obtained in two independent reactions. b, Co-immunoprecipitation of endogenous MITOK using mitochondria isolated from HeLa cells. FT, flow-through fraction; W3, third (last) co-immunoprecipitation wash. Representative of two independent experiments. c, Co-immunoprecipitation between overexpressed mouse MITOK and mutant human MITOSUR(K513A). Representative of two independent experiments.
Extended Data Fig. 7 Genetic ablation of MITOK in HeLa cells.
a, Schematic of the MITOK gene. The expanded regions were used to design Cas9 guides (highlighted in red). b, Western blot of wild-type and MITOK-knockout HeLa cell lines. Representative of three independent experiments. c, Mitochondrial morphology in wild-type and MITOK-knockout HeLa cells. Scale bar, 10 μm. Asterisks are located near doughnut-shaped mitochondria. Similar results were obtained in five independent experiments. d, e, ΔΨm measurements in control and MITOK-knockout cells. Cells were loaded with TMRM, and normalized fluorescence in different regions was monitored through time. d, Representative traces of single mitochondria. e, Pseudo-coloured representative images of a HeLa cell knockout for MITOK, loaded with TMRM at the indicated time points. Similar results were obtained in four independent experiments. f, Western blot in HeLa cells of the indicated genotype. Representative of two independent experiments.
Extended Data Fig. 8 Loss of MITOK causes mitochondrial dysfunction.
a, OCR measurements in wild-type and MITOK-knockout HeLa cells treated with either vehicle or 1 pM valinomycin for 1 h. Representative of three independent experiments. b, OCR measurements in wild-type and MITOK-knockout HeLa cells transfected with control or mitoKATP-expressing (MITOSUR-P2A-MITOK) plasmids. Representative of three independent experiments. c, Maximal cristae width in the indicated genotype. n ≥ 12 individual cells (approximately 20 cristae per cell were measured) from 2 independent preparations. *P ≤ 0.013 using two-way ANOVA with Holm–Sidak correction. d, OPA1 crosslinking (using 1 mM BMH) in wild-type and MITOK-knockout cells. Similar results were obtained in three independent experiments. e, f, Extracellular acidification rate (ECAR) (e) and OCR (f) measurements in intact cells of the indicated genotype. n = 5 biological replicates, representative of 2 independent experiments. g, ROS production during energy stress. Cells were incubated in 5.5 mM of either glucose or 2-deoxyglucose in the presence or absence of 30 μM diazoxide, and fluorescence was monitored for 16 h. Box plots indicate the rate of ROS production over this time frame. n ≥ 10 biological replicates from 3 independent experiments. *P < 0.05 using three-way ANOVA with Holm–Sidak correction. h, Cell death analysis in HeLa cells treated with 0, 100 or 500 μM H2O2. Data are normalized to the untreated condition, and expressed as mean ± s.d. n = 3 independent experiments. *P < 0.003 using two-way ANOVA with Holm–Sidak correction.
Supplementary information
Supplementary Information
Supplementary figure 1: contains uncropped images of Western blot membranes used for the assembly of the indicated figures.
Rights and permissions
About this article
Cite this article
Paggio, A., Checchetto, V., Campo, A. et al. Identification of an ATP-sensitive potassium channel in mitochondria. Nature 572, 609–613 (2019). https://doi.org/10.1038/s41586-019-1498-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1498-3
This article is cited by
-
The mitochondrial ATP-dependent potassium channel (mitoKATP) controls skeletal muscle structure and function
Cell Death & Disease (2024)
-
Insulin Secretion and the β-Cell 102 Years After the Discovery of the Hormone
Current Molecular Biology Reports (2024)
-
Nicorandil Exerts Anticonvulsant Effects in Pentylenetetrazol-Induced Seizures and Maximal-Electroshock-Induced Seizures by Downregulating Excitability in Hippocampal Pyramidal Neurons
Neurochemical Research (2023)
-
Loss of the large conductance calcium-activated potassium channel causes an increase in mitochondrial reactive oxygen species in glioblastoma cells
Pflügers Archiv - European Journal of Physiology (2023)
-
Mitochondrial connexin43 and mitochondrial KATP channels modulate triggered arrhythmias in mouse ventricular muscle
Pflügers Archiv - European Journal of Physiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.