Abstract
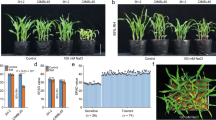
The size of plants is largely determined by growth of the stem. Stem elongation is stimulated by gibberellic acid1,2,3. Here we show that internode stem elongation in rice is regulated antagonistically by an ‘accelerator’ and a ‘decelerator’ in concert with gibberellic acid. Expression of a gene we name ACCELERATOR OF INTERNODE ELONGATION 1 (ACE1), which encodes a protein of unknown function, confers cells of the intercalary meristematic region with the competence for cell division, leading to internode elongation in the presence of gibberellic acid. By contrast, upregulation of DECELERATOR OF INTERNODE ELONGATION 1 (DEC1), which encodes a zinc-finger transcription factor, suppresses internode elongation, whereas downregulation of DEC1 allows internode elongation. We also show that the mechanism of internode elongation that is mediated by ACE1 and DEC1 is conserved in the Gramineae family. Furthermore, an analysis of genetic diversity suggests that mutations in ACE1 and DEC1 have historically contributed to the selection of shorter plants in domesticated populations of rice to increase their resistance to lodging, and of taller plants in wild species of rice for adaptation to growth in deep water. Our identification of these antagonistic regulatory factors enhances our understanding of the gibberellic acid response as an additional mechanism that regulates internode elongation and environmental fitness, beyond biosynthesis and gibberellic acid signal transduction.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The gene sequences used in this work have been deposited at DDBJ (https://www.ddbj.nig.ac.jp/index-e.html; ACE1C9285: LC543529; ACE1 short transcript: LC543530; and DEC1C9285: LC543531). The RNA-sequencing datasets analysed during this study are available in a previous study46. Microarray data for trans-eQTL analysis are available in the NCBI Gene Expression Omnibus (GEO) database (accession number GSE87702). The fastq files of O. rufipogon are available in OryzaGenome (from the ‘Deep Sequenced Accessions List’; http://viewer.shigen.info/oryzagenome2detail/index.xhtml). The 3K RG 29mio biallelic SNPs dataset is registered in the 3K RGP and OryzaSNP project (IRRI, http://snp-seek.irri.org/download.zul). Source data are provided with this paper.
References
Kaneko, M. et al. Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35, 104–115 (2003).
Kende, H., van de Knaap, E. & Cho, H.-T. Deepwater rice: a model plant to study stem elongation. Plant Physiol. 118, 1105–1110 (1998).
Sauter, M., Mekhedov, S. L. & Kende, H. Gibberellin promotes histone H1 kinase activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deepwater rice internodes. Plant J. 7, 623–632 (1995).
Cronk, Q. C. B. The Molecular Organography of Plants (Oxford Univ. Press, 2009).
Sasaki, A. et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416, 701–702 (2002).
Peng, J. et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261 (1999).
Catling, D. Rice in Deep Water (Palgrave Macmillan, 1992).
Kuroha, T. et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 361, 181–186 (2018).
Kawahara, H., Chonan, N. & Wada, K. Studies on morphogenesis in rice plants. 3. Interrelation of the growth among leaves, panicle and internodes, and a histological observation on the meristem of culm. Proc. Crop Sci. Soc. Jpn 37, 372–383 (1968).
Suetsugu, I. Studies on the first jointing stage in rice plants. Proc. Crop Sci. Soc. Jpn 37, 489–498 (1968).
Inouye, J. On floating rice and its ecological traits in Southeast Asia. Southeast Asian Studies 25, 51–61 (1987).
Hedden, P. & Thomas, S. G. (eds) Annual Plant Reviews: the Gibberellins (Wiley-Blackwell, 2016).
Nagai, K. et al. QTL analysis of internode elongation in response to gibberellin in deepwater rice. AoB Plants 6, plu028 (2014).
Hattori, Y. et al. A major QTL confers rapid internode elongation in response to water rise in deepwater rice. Breed. Sci. 57, 305–314 (2007).
Inouye, J., Kyuragi, T. & Xuan, V.-T. On the growth habits of floating, single- and double-transplanted rice plants in the Mekong Delta. Japan. J. Trop. Agr. 22, 67–70 (1978).
Kotogány, E., Dudits, D., Horváth, G. V. & Ayaydin, F. A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6, 5 (2010).
Kania, T., Russenberger, D., Peng, S., Apel, K. & Melzer, S. FPF1 promotes flowering in Arabidopsis. Plant Cell 9, 1327–1338 (1997).
Sakamoto, T. et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134, 1642–1653 (2004).
Hattori, Y. et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030 (2009).
Gómez-Ariza, J. et al. A transcription factor coordinating internode elongation and photoperiodic signals in rice. Nat. Plants 5, 358–362 (2019).
Pauwels, L. et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 (2010).
Li, S. et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl Acad. Sci. USA 110, 3167–3172 (2013).
Kojima, M. et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50, 1201–1214 (2009).
Shinozaki, Y. et al. Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 83, 237–251 (2015).
Minami, A. et al. Cold acclimation in bryophytes: low-temperature-induced freezing tolerance in Physcomitrella patens is associated with increases in expression levels of stress-related genes but not with increase in level of endogenous abscisic acid. Planta 220, 414–423 (2005).
Ueguchi-Tanaka, M. et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19, 2140–2155 (2007).
Zhang, H.-B., Zhao, X., Ding, X., Paterson, A. H. & Wing, R. A. Preparation of megabase-size DNA from plant nuclei. Plant J. 7, 175–184 (1995).
International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature 436, 793–800 (2005).
Mikami, M., Toki, S. & Endo, M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 88, 561–572 (2015).
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994).
Kuroha, T. et al. eQTLs regulating transcript variations associated with rapid internode elongation in deepwater rice. Front. Plant Sci. 8, 1753 (2017).
Arends, D., Prins, P., Jansen, R. C. & Broman, K. W. R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26, 2990–2992 (2010).
Cubillos, F. A. et al. Expression variation in connected recombinant populations of Arabidopsis thaliana highlights distinct transcriptome architectures. BMC Genomics 13, 117 (2012).
Yamaji, N. & Ma, J. F. Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 143, 1306–1313 (2007).
Himuro, Y. et al. Arabidopsis galactinol synthase AtGolS2 improves drought tolerance in the monocot model Brachypodium distachyon. J. Plant Physiol. 171, 1127–1131 (2014).
Alves, S. C. et al. A protocol for Agrobacterium-mediated transformation of Brachypodium distachyon community standard line Bd21. Nat. Protocols 4, 638–649 (2009).
Hisano, H. & Sato, K. Genomic regions responsible for amenability to Agrobacterium-mediated transformation in barley. Sci. Rep. 6, 37505 (2016).
Takahashi, W. & Takamizo, T. Plant regeneration from embryogenic calli of the wild sugarcane (Saccharum spontaneum L.) clone ‘Glagah Kloet ’. Bull. NARO Inst. Livest. Grassl. Sci. 13, 23–32 (2013).
Takahashi, W., Oishi, H., Ebina, M., Takamizo, T. & Komatsu, T. Production of transgenic italian ryegrass (Lolium multiflorum Lam.) via microprojectile bombardment of embryogenic calli. Plant Biotechnol. 19, 241–249 (2002).
Spangenberg, G. et al. Transgenic tall fescue (Festuca arundinacea) and red fescue (F. rubra) plants from microprojectile bombardment of embryogenic suspension cells. J. Plant Physiol. 145, 693–701 (1995).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Browning, B. L. & Browning, S. R. Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 98, 116–126 (2016).
Szpiech, Z. A. & Hernandez, R. D. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 31, 2824–2827 (2014).
Sabeti, P. C. et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837 (2002).
Voight, B. F., Kudaravalli, S., Wen, X. & Pritchard, J. K. A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006).
Minami, A. et al. Time-course transcriptomics analysis reveals key responses of submerged deepwater rice to flooding. Plant Physiol. 176, 3081–3102 (2018).
Taoka, K. et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335 (2011).
Kobayashi, K. et al. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24, 1848–1859 (2012).
Tsuji, H. et al. Hd3a promotes lateral branching in rice. Plant J. 82, 256–266 (2015).
Kaneko-Suzuki, M. et al. TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol. 59, 458–468 (2018).
Ikeda, A. et al. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010 (2001).
Fu, X. et al. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14, 3191–3200 (2002).
Hoffmann-Benning, S. & Kende, H. On the role of abscisic acid and gibberellin in the regulation of growth in rice. Plant Physiol. 99, 1156–1161 (1992).
Azuma, T., Mihara, F., Uchida, N., Yasuda, T. & Yamaguchi, T. Plant hormonal regulation of internodal elongation of floating rice stem sections. Japan. J. Trop. Agr. 34, 271–275 (1990).
Nemoto, K., Ukai, Y., Tang, D.-Q., Kasai, Y. & Morita, M. Inheritance of early elongation ability in floating rice revealed by diallel and QTL analyses. Theor. Appl. Genet. 109, 42–47 (2004).
Tang, D.-Q., Kasai, Y., Miyamoto, N., Ukai, Y. & Nemoto, K. Comparison of QTLs for early elongation ability between two floating rice cultivars with a different phylogenetic origin. Breed. Sci. 55, 1–5 (2005).
Kawano, R., Doi, K., Yasui, H., Mochizuki, T. & Yoshimura, A. Mapping of QTLs for floating ability in rice. Breed. Sci. 58, 47–53 (2008).
Han, Y. et al. Overexpression of OsSIN, encoding a novel small protein, causes short internodes in Oryza sativa. Plant Sci. 169, 487–495 (2005).
Ohtsubo, Y., Ikeda-Ohtsubo, W., Nagata, Y. & Tsuda, M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9, 376 (2008).
Sauter, M. & Kende, H. Gibberellin-induced growth and regulation of the cell division cycle in deepwater rice. Planta 188, 362–368 (1992).
Ge, L. et al. Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol. 135, 1502–1513 (2004).
Acknowledgements
We thank H. Morishima (deceased 2010) for encouragement to do this work and K. Suzuki, Y. Kondo, M. Koike, A. Minami, S. Reuscher, N. Yamaji, J. F. Ma and S. Sasaki for technical assistance; M. Mikami, S. Toki and M. Endo (NARO) for providing the CRISPR–Cas9 vectors; M. Ueguchi-Tanaka for providing the SLR1 antibody; and R. B. Angeles-Shim for English language editing. Deepwater rice, wild rice accessions and barley cv. Golden Promise were provided by the National Bioresource Project (NBRP) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. This work was supported by the CREST programme of the Japan Science and Technology Agency (JST) (JPMJCR13B1); by the SATREPS programme (JPMJSA1706) of the JST and Japan International Cooperation Agency (JICA); by MEXT/JSPS KAKENHI (16K18565, 16H06466, 17H06473 and 19K15815); and by Genomics for Agricultural Innovation (QTL5003) of the Ministry of Agriculture, Forestry, and Fisheries of Japan (MAFF). This study was supported in part by RIKEN-Nagoya University Science and Technology Hub.
Author information
Authors and Affiliations
Contributions
K.N. and M.A. designed the study. K.N. and S.I. carried out the genetic linkage analysis. J.W. and T.F. performed the screening of BAC clones and sequence analysis. K.N., R.G., Y.N., M.F., Y.H., M. Kobayashi, W.A., H.H. and K.S. performed the transgenic experiments and sequence analysis. K.N., Y.M., R.G., T.H., A.Y. and H.T. elucidated the molecular mechanisms. M. Kojima, Y.T. and H.S. quantified hormone levels. A.F. performed the trans-eQTL analysis. Y.S. and T.A. performed the evolutionary analyses. K.N., T.A. and M.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Patrick Achard, Julia Bailey-Serres and Bin Han for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Schematic of a rice plant and the concept of timing of internode elongation.
a–c, Measurement and concept of initiation of internode elongation. The growth stage of rice plant is defined by the number of fully expanded leaves. For example, the right plant in a is at the 6-leaf stage. Plant height and total internode length were defined as the length from the base to the leaf top and from the base to the end of the uppermost internode, respectively. a, Schematic of rice showing no internode elongation during the vegetative phase. b, Schematic of rice showing internode elongation at the 6-leaf stage. c, Schematic of rice showing internode elongation at the 3-leaf stage. It has been reported that the lowest elongated internode (LEI), proposed as an index of initiation of internode elongation, is correlated with the total internode length and number of elongated internodes. In a, LEI was not determined because the plant did not elongate internodes. By contrast, the LEI was 6 and 3 because internode elongation started from the 6- and 3-leaf stages in b and c, respectively. Initiation of internode elongation is influenced by activation of the intercalary meristem in internodes3 and comparison of LEI reveals the timing of the acquisition of internode elongation ability. However, the factors that directly control this trait have not yet been identified.
Extended Data Fig. 2 Changes in shoot phenotype in shallow-water and deep-water conditions.
a, The plant height of T65 and C9285 was measured at each leaf stage in shallow-water conditions. Data are mean ± s.d. (n ≥ 3 plants, Source Data). b, Expression level of OsMADS14 (OsAP1B: LOC_Os03g54160) and OsMADS15 (OsAP1A: LOC_Os07g01820) at each growth stage of T65 (n = 3 plants) and C9285 (left: n = 3 plants; right: n = 4 plants). Data are mean ± s.d. One-way ANOVA followed by Tukey’s multiple-comparison test. OsMADS14 and OsMADS15 are activated during the growth phase transition from the shoot apical meristem to the inflorescence meristem by Hd3a (rice FT), OsFD1 and 14-3-3 protein complex47,48,49,50. Therefore, OsMADS14 and OsMADS15 were used as phase transition markers. RNA was extracted from shoot apices at the 5-, 7- and 12-leaf stages containing shoot apical meristem, base of young leaves, immature internodes and nodes. OsMADS14 and OsMADS15 expression was low at the 7-leaf stage but their expression drastically increased at the 12-leaf stage in T65, suggesting that the phase transition occurred at this stage in natural conditions. Internode elongation started after phase transition (T65 elongated internodes after the 11-leaf stage (Fig. 1c)). By contrast, C9285 did not express OsMADS14 and OsMADS15 at these leaf stages. Right panels, expression level of OsMADS14 and OsMADS15 in 1- and 7-month plants of deepwater rice C9285. C9285 showed expression of OsMADS14 and OsMADS15 3 months later than normal paddy rice. This suggests that C9285 elongates internodes without a phase transition (Fig. 1c). c–e, Plant height (c), total internode length (d) and number of elongated internodes (e) after seed maturation. Data are mean ± s.d. (n = 5 plants). Numbers indicate P value derived from a two-tailed t-test. T65 required 6 months from germination until seed maturation whereas C9285 needed 9 months in our condition. Numbers above bars indicate significant differences by Student’s t-test. f, Hormone contents during plant growth in shallow-water and deep-water conditions. GA1, GA4, abscisic acid (ABA) and indoleacetic acid (IAA) contents at the 5-leaf stage (top) and 8-leaf stage (bottom) of T65 and C9285 in shallow-water and deep-water conditions. Data are mean ± s.d. (n = 3 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. g, Temporal responses of internode length to 10−4 M GA. GA treatment was started from germination and internode length was measured weekly. Data are mean ± s.d. (n = 6 plants). Numbers indicate P values derived from a two-tailed t-test compared with C9285 and T65 for each point. h, Internode length of T65 and C9285 according to GA concentration in Fig. 1g. Right, an enlarged view, from 0 to 2 cm, of the left panel. Colours show internode lengths. Data are mean ± s.d. The number of plants (n) is shown. i, SLR1 degradation after GA treatment. Western blotting of degradation of SLR1 after GA treatment using a rabbit anti-SLR1 antibody. SLR1, a DELLA protein of rice and master growth repressor in GA signalling, is degraded by the 26S proteasome through the GA-dependent SCFGID1 complex, resulting in derepression of GA responses51,52. GA treatment was performed on 6-LS plants of T65 and C9285, and total proteins of the stem section were extracted. Arrow indicates the position of SLR1. Ponceau-S, loading control. GA-dependent degradation of SLR1 was observed in the internode of T65, which showed no internode elongation by GA, as in C9285. All experiments except hormone quantification (one time) were repeated twice with similar results.
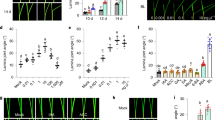
Extended Data Fig. 3 Internode elongation with growth phase and QTLs related to GA and the deep-water response.
a, Normal paddy rice does not elongate internodes during the vegetative phase, but it elongates internodes immediately after the reproductive phase (attributed to promotion of GA biosynthesis) in shallow-water conditions. In deep-water conditions, internodes do not elongate during the vegetative phase8,14. Therefore, the plant cannot survive owing to oxygen deprivation in deep-water conditions. b, Deepwater rice shows vigorous growth and an increased plant height owing to internode elongation even in shallow-water conditions (Fig. 1c, Extended Data Fig. 2). Furthermore, when deepwater rice is exposed to deep-water conditions, it exhibits rapid internode elongation owing to GA accumulation, resulting in exposure of leaves on the water, thus avoiding oxygen deficiency8. c, Regulation of plant hormone crosstalk induced by deep water during the vegetative phase. The deep-water condition induces ethylene accumulation in the plant body in deepwater rice and normal paddy rice, which reduces the level of abscisic acid, an antagonist of GA19,53,54. Accumulated ethylene also increases the expression level of GA20OX2, which is involved in GA biosynthesis, in deepwater rice. However, accumulated ethylene does not stimulate GA20OX2 expression in T658. Therefore, internode elongation by GA is induced in deepwater rice but not in normal paddy rice. d, QTL analysis of GA-responsive internode elongation using recombinant inbred lines of normal paddy rice (T65) and deepwater rice (Bhadua) detected five QTLs on chromosome 3, 8, 9, 10 and 12 (red)13. QTLs for total internode length (TIL) in deep-water conditions were detected on chromosomes 1 and 12 (blue)14. The positions of QTLs are shown with reference to the physical distance of the markers in the QTL analyses. The gene responsible for the QTL on chromosome 1 was GA20OX2, which encodes a GA-synthesis enzyme8. We also identified two ethylene-related transcription-factor genes, SK1 and SK2, as responsible for the QTLs of TIL on chromosome 1219. The QTL regulating the lowest elongated internode (LEI) that regulates the timing of initiation of internode elongation in deep-water conditions was detected on chromosomes 3 and 12 (orange)14,55,56,57. The QTLs for GA response on chromosomes 3 and 12 and the LEI for the deep-water response overlapped. Therefore, the same genes may be responsible for these QTLs. e, Genotypes of near-isogenic lines (NILs). Red and blue, C9285 and T65 genomic regions, respectively. NIL3 and NIL12 possess the QTL regions on chromosomes 3 and 12 for the initiation of internode elongation in response to GA and deep-water conditions, respectively, in T65 genetic background. NIL3+12 possesses both QTLs. f, g, Total internode length of T65, NIL1, NIL3+12 and NIL1+3+12 after exogenous GA treatment or in deep-water conditions. f, Total internode length after mock and exogenous GA treatment for 3 weeks from germination. Number indicates P value derived from a two-tailed t-test (mean ± s.d., n = 9 plants). g, Total internode length of 8-LS plants in shallow-water and deep-water conditions for 1 week. Number indicates P value derived from a two-tailed t-test (mean ± s.d., n = 4 plants). h, Mapping of QTLs on chromosome 3. NIL3+12 and NIL12 were used as the parental lines (P). Red and blue, C9285 and T65 genomic regions, respectively. NIL3+12 has QTLs on chromosomes 3 and 12, whereas NIL12 has a QTL on chromosome 12. The F1 population derived from crossing of parental lines resulted in a heterozygous genotype on chromosome 3 and a C9285 homozygous genotype on chromosome 12. By selfing of F1, an F2 recombinant population with segregating genotypes on chromosome 3 was produced. F2 plants showing recombination between the two markers (RM7249 and 9k were used for initial screening) were selected (red dotted rectangles). The F3 population was produced by selfing of the F2 plants. The F3 population was expected to be segregated into three genotypes: homozygous with recombination, heterozygous, and homozygous with no recombination in a 1:2:1 ratio. Homozygotes were selected and used for high-resolution linkage analysis with GA treatment for 3 weeks after germination. For positional cloning of QTL on chromosome 12, the mapping population derived from the cross between NIL3+12 and NIL3 was used, and subsequent genotyping was performed using DNA markers on chromosome 12 (Supplementary Table 5). i, j, Dominant effect of the QTL on chromosome 3 on the response to GA. i, Genotype of F1 derived from the cross between NIL3+12 and NIL12. Only chromosome 3 showed a heterozygous genotype and segregated in F2 progeny. j, The segregating genotype (left) and internode length (right). Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. The progenies of #59 and #58 plants segregated as C9285-type homozygote, T65-type homozygote, and heterozygote at the region of RM7249 to 9k. Heterozygous plants showed similar internode length to C9285-type homozygotes, suggesting that the C9285-type QTL on chromosome 3 exerts the dominant effect on the GA response of internodes. k, Comparison of internode elongation in response to GA and deep-water conditions. Left, genotype of the GA-mapping lines. Middle, internode length of the mapping lines with GA treatment for 3 weeks. Right, internode length of mapping lines in deep-water conditions for 1 week. Data are mean ± s.d. The number of plants (n) is shown. Number indicates P value derived from a two-tailed t-test. l, Internode length of T65 and NIL3 in shallow-water or deep-water conditions using 8-leaf-stage plants. Deep-water conditions were maintained for 1 week. Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. The data suggest the QTL on chromosome 3 that regulates initiation of internode elongation (LEI for the deep-water response) corresponds to the GA-responsive QTL. All experiments were repeated twice with similar results.
Extended Data Fig. 4 The role of ACE1.
a, Coding sequence of ACE1. The ACE1 in T65 is 321 bp in size and that of C9285 is 327 bp. ACE1 of T65 has an additional guanine (G) and one single-nucleotide polymorphism at the end of the coding sequence. b, Alignment of the amino acid sequences of ACE1. Nuclear localization signal (NLS) was predicted in ACE1T65, but not in ACE1C9285 by WOLF PSORT (https://www.genscript.com/wolf-psort.html). c, Structure of ACE1. The short ACE1 transcript in T65 and C9285 is 207 bp. Grey and blue boxes, non-coding and coding sequences, respectively. Hatched box, region encoding different amino acids compared with other sequences. The allele for the long ACE1 transcript in T65 (ACE1T65) has been reported as OsSIN (Oryza sativa SHORT INTERNODE)58. OsSIN has a 1-bp insertion at the same location as ACE1T65. Ref. 58 reported that constitutive expression of OsSIN (ACE1T65) indicated dwarfism, but we did not observe a dwarf phenotype in T65 background. This may be due to differences in the genetic background. d, Amino acid sequences of long and short ACE1 proteins in T65 and C9285. The short ACE1 protein was identical in T65 and C9285. e, Gross morphology of plants that express ACE1T65 or ACE1C9285 in T65 genetic background (T1) after mock and GA treatment. Transgenic plants were treated with 10−4 M GA for 3 weeks after germination. Scale bars, 5 cm. Arrowheads, nodes linked by elongated internodes. f, Quantitative data of the total internode length of plants in e. Data are mean ± s.d. (n = 10 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. g, Quantification of OsMADS14 and OsMADS15 expression in control and ACE1C9285-overexpressing plants (T1) after mock or GA treatment. Data are mean ± s.d. (n = 4 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. T65 at the vegetative (5-LS) and reproductive (12-LS) phases were used as controls. GA was applied to 6-leaf-stage plants for 1 week. RNA was extracted from an approximately 1 cm region of the shoot apex including the shoot apical meristem, developing leaves and internodes. Because the ACE1C9285-overexpressing plants did not express OsMADS14 and OsMADS15, they were in the vegetative phase. h, Gross morphology of ACEC9285ox plants and plants that overexpress the short ACE1 transcript (ACE1shortox) in the T65 genetic background (T0) after mock and GA treatment. Right upper panel shows enlarged view of base of the plant. i, Quantitative data of the total internode length of plants in h. GA was applied for 2 weeks. Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. j, Total internode length of plants that overexpress ACE1 in the NIL12 genetic background (T0) without GA treatment. Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. k, Gross morphology (top panels) and enlarged view (bottom panels) of the vector control plants and those that overexpress ACE1C9285 in the NIL1+3+12 genetic background. Scale bars, 10 cm. l, m, Total internode length (l) and number of elongated internodes (m) of plants in k. Data are mean ± s.d. The number of plants (n) is shown. Numbers indicate P values derived from a two-tailed t-test. n, Gross morphology (left) and enlarged view (right) of plants that overexpress ACE1C9285 (T0) in C9285 genetic background. Arrowheads indicate nodes linked by elongated internodes. Scale bars, 10 cm. The creeping-like stem elongation is likely to be due to rapid stem elongation that cannot support shoot weight; therefore, internodes bend and touch the ground. All experiments were repeated twice with similar results.
Extended Data Fig. 5 Quantification of ACE1 expression in plant tissues.
a, Schematic of a rice plant showing the organs from which RNA was extracted to assess ACE1 expression. Total RNA was extracted from roots, leaf sheath (LS), youngest fully expanded leaf blade (LB), elongating leaf blade (ELB), shoot apex (SA) and elongating internode. The internode was sampled separately at the base (lower) and middle part of the internode (middle). b, Quantification of ACE1 expression in shallow-water and deep-water conditions. Deep-water conditions were maintained for 1 week. Data are mean ± s.d. (n = 3 plants). Numbers indicate P values derived from a two-tailed t-test. c–f, Quantification of expression of cell-elongation-related genes, OsEXPA4 and OsEXPB4, and cell-division-related genes, CDKA1 and CYCLINB2;1, in internodes in shallow-water or deep-water conditions. Data are mean ± s.d. (n = 3 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. Cell-elongation-related genes showed higher expression in the middle region whereas cell-division-related genes showed higher expression in the lower region. These results suggest ACE1 is preferentially expressed in the cell-elongation zone (middle region) rather than the cell-division zone (lower region). Deep-water treatment was conducted for one week. All experiments were repeated twice with similar results.
Extended Data Fig. 6 Screening of transactivating factors of the ACE1 promoter.
a, Trans-eQTL region of ACE1 on chromosome 9. The genome-wide eQTL mapping identified 7,145 significant trans-eQTLs (false discovery rate < 0.05) using the R/qtl32 and R/eqtl packages33. The genotype and expression data were derived from a previous study31 (85 individuals of the T65/Bhadua RILs). Of these, we detected one trans-regulatory region on chromosome 9 as a trans-eQTL (limit of detection, 2.5; maximum limit of detection peak marker, ad09003568) that regulates the expression of ACE1 (Os03g0346200). In this analysis, we used a support interval of 1.5 and the default settings in the R/eqtl::define.peak() function to evaluate QTLs and support intervals. b, Expression of transcription-factor genes in the trans-eQTL region. Among the genes included in the overlapping region of trans-eQTL and GA-related QTL analyses, eight transcription-factor genes were selected from the RNA sequence data of a previous study46. Data are mean ± s.d. (n = 3 plants). The expression of Os09t0306400 (bZIP71), Os09g0287000 (SUB1B) and Os09g0286600 (SUB1C) was altered in deep-water conditions. These genes were used as effectors in the transactivation analysis. c, About 4.7 kb upstream of the ACE1T65-coding region and 4.4 kb upstream of the ACE1C9285-coding region were amplified from genomic DNA of T65 and C9285 by PCR, respectively, and fused with hRluc (R. reniformis) in pUC19. These constructs were used as reporters in the transactivation analysis. Black lines and colored charts indicate genomic sequences and sequence homologies, respectively. Sequence alignment was constructed by GenomeMatcher59. d, Screening of transactivating factors of the ACE1 promoter using rice seeds. Constructs were introduced by bombardment. Data are mean ± s.d. (n = 3 experiments). Numbers indicate P values derived from a two-tailed t-test when compared with VC. e, Effect of SUB1C overexpression on ACE1 expression in T65 (left) and NIL3 (right). NIL3 possesses ACE1C9285 in a T65 (Extended Data Fig. 3e). X-axis, expression level of SUB1C; y-axis, expression level of ACE1. Dots indicate the expression levels of individual plants (n = 6 plants). R2, logarithmic approximation. f, Subcellular localization of SUB1C–GFP in onion epidermal cells. 35S:mCherry was used as a control. Fluorescence profiles were imaged by the profile tool of the ZEN software (Carl Zeiss). Plasmid constructs were introduced by particle bombardment. g, Transactivation assay of ACE1 promoter by SK1 and SK2. Data are mean ± s.d. (n = 3 experiments). Numbers indicate P values derived from a two-tailed t-test when compared with VC. h, ACE1T65–GFP and ACE1C9285–GFP in onion epidermal cells. 35S:mCherry was used as a control. Fluorescence profiles were imaged by the profile tool of the ZEN software (Carl Zeiss). Plasmid constructs were introduced by particle bombardment. Nuclear localization signal was predicted in ACE1T65, but not in ACE1C9285 by WOLF PSORT (https://www.genscript.com/wolf-psort.html). The GFP fluorescence signal of 35S:GFP and 35S:ACE1C9285-GFP overlapped with the mCherry signal, whereas 35S:ACE1T65-GFP showed a high intensity of GFP signal in the nucleus, suggesting that ACE1T65 is preferentially localized in the nucleus. All experiments were repeated twice with similar results.
Extended Data Fig. 7 Tissue localization of ACE1 after GA treatment or in deep-water conditions.
a, Validation of an antibody against ACE1. Western-blot analysis using anti-GST (left) and anti-ACE1 (middle) antibodies after SDS–PAGE of crude protein extracts from E. coli including GST-ACE1 inducing construct. GST-ACE1 protein producing was induced by adding 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). Arrow indicates the position of GST–ACE1. CBB staining, loading control (right). b, Schematic of internode. c, Immunostaining of ACE1 (red) in C9285 after GA treatment for 10 days. d, e, Enlarged views of red and yellow dashed squares in c. f–i, Tissue localization of ACE1 in shallow-water and deep-water conditions. Deep-water treatment was conducted for one day (g) and 10 days (h), and compared with shallow-water treatment (f). i, Enlarged view of red dashed square in h. ACE1 signal was localized throughout the internode, except up to about 2 mm above the node, in early stage, and subsequently, it was detected from about 2–5 mm above the node 10 days later, whereas it was hardly detected about 10 mm above from node (d, h). Scale bars, 1 mm. All experiments were repeated three times with similar results.
Extended Data Fig. 8 ACE1 function relates to intercalary meristem activity in concert with GA.
a, Schematic of internodes from which histological observations and mRNA expression analysis were conducted in b–e. b, c, EdU signal (green dots) at the shoot apex (b) and lower part (c) of an elongating internode of C9285 in shallow-water conditions. d, Enlarged view of red dashed square in c. e, Expression of cell-division-related genes, HISTONE H4, CDKA1 and CYCLINB2;1, at the shoot apex and upper and lower elongating internodes in shallow-water conditions. Data are mean ± s.d. (n = 3 plants). Numbers indicate P values derived from a two-tailed t-test when compared with upper and lower. The EdU-positive region matches the area of expression of cell-division-related genes. f, Visualization of cell-division zone in VC and ACE1C9285ox by EdU. The sixth internodes of 5-LS plants are highlighted (red arrows). 10−4 M GA treatment was conducted for 0, 3, 5 and 10 days. Scale bars, 1 mm. g, Time-dependent changes in internode length of VC and ACE1C9285ox in T65 genetic background in response to GA treatment. Right, enlarged view of the data from 0 to 5 days. Data are mean ± s.d. The number of plants (n) is shown. Numbers indicate P values derived from a two-tailed t-test when compared with VC. All experiments were repeated three times with similar results. h, Model of internode GA response in the absence of ACE1. Cell division occurs during internode development without ACE1. However, competency for the GA response cannot be acquired by this internode, hence, the intercalary meristem does not develop or meristem activity may not be continued, and internodes are differentiated, in the presence of GA. i, Model of internode GA response in the presence of ACE1. ACE1 confers competence for GA response in the internode to develop intercalary meristem. Subsequently, intercalary meristem activity and its maintenance in internodes increases in concert with GA, resulting in the initiation of internode elongation. Notably, although ACE1C9285 expression was highest at the cell-elongation zone, 10–30 mm from the node (Extended Data Fig. 5b), ACE1C9285 protein was detected in the region 2–5 mm from the node after GA treatment and deep-water conditions for 10 days (Fig. 3a–d, Extended Data Fig. 7) which coincided with the region in which the EdU signal was detected (Extended Data Fig. 8). In addition, overexpression of ACE1C9285 induced cell division in coordination with GA in the basal region of elongating internode (Fig. 3e). These results suggest that the protein stability of ACE1C9285 may increase at intercalary meristem, or mRNA or protein is transported and accumulated in this region to control intercalary meristem activity. It has previously been proposed that the cell cycle entry at intercalary meristem might be a consequence of GA-induced cell elongation, as cell elongation of deepwater rice internode occurred before cell division with GA treatment3,60. The high expression of ACE1 in the cell-elongation zone above intercalary meristem may suggest that ACE1 is a regulatory molecule that controls cell division by transmitting cell-elongation signals to intercalary meristem in GA-dependent manner. Further verifications such as changes in subcellular localization by GA treatment, RNA or protein stability or mobility, and identification of downstream targets of ACE1 are required to understand the mechanism of the molecular function.
Extended Data Fig. 9 Role of ACL1 in normal paddy rice.
a, Phylogenetic tree constructed using the full-length amino acid sequences of ACE1 homologues from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) using MEGA 7. Numbers on branches are the 1,000-bootstrap values. b, The model of ACE1C9285 protein sequence. The numbers indicate the amino acid positions. ACE1C9285 and homologues possess three motifs of unknown function (Supplementary Table 2). c–e, A phylogenetic tree of ACE1 homologues in the mosses, pteridophytes (c), dicotyledon subgroup I clade (d) and dicotyledon subgroup II clade (e) in a. Dicotyledon subgroup I contains FPF117, an Arabidopsis homologue of ACE1. Dicotyledonous and monocotyledonous ACE1 homologues formed individual clades without mixing. Numbers on branches are the 1,000-bootstrap values. f, Overexpression of Arabidopsis FPF1 in T65 genetic background (T0). Top, gross morphology of the vector control plants and plants that overexpress pACE1C9285 or FPF1 in the T65 genetic background. FPF1 was cloned from a Col-0 plant. GA treatment was performed for 2 weeks. Arrowheads indicate nodes linked by elongated internodes. Scale bars, 1 cm. Bottom, total internode length. Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. Arabidopsis FPF1 was overexpressed in rice; however, it did not induce internode elongation, suggesting that ACE1 and FPF1 have distinct functions in rice and Arabidopsis, respectively. g, Sequence alignment of ACE1 homologues in rice. Among the six rice homologues, ACE1 had the highest homology with LOC_Os07g47450, and we named it ACE1-LIKE 1 (ACL1). Although LOC_Os01g15340 has been termed ROOT ARCHITECTURE ASSOCIATED1 (RAA1)61, its involvement in internode elongation has not been reported. h, Quantification of OsMADS14, OsMADS15 and ACL1 expression at each leaf stage in Nipponbare in shallow-water conditions. Data are mean ± s.d. (n = 4 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. i, Sequence of the acl1 mutant generated using the CRISPR/Cas9 system. j, Gross morphology of the wild-type (WT) and the acl1 mutants at mature stage. Scale bars, 1 m. k, Quantitative data of j. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. All experiments were repeated twice with similar results.
Extended Data Fig. 10 The role of ACE1 in Gramineae.
a, Gross morphology of ACE1C9285-overexpressing plants in B. distachyon without GA treatment after ripening. Scale bars, 5cm. b, Quantification of ACE1 expression in ACE1-overexpressing B. distachyon plants. Data are mean ± s.d. (n = 4 plants). Numbers indicate P values derived from a two-tailed t-test when compared with VC. c, d, Total internode length of ACE1-overexpressing B. distachyon plants after mock (c) or GA treatment (d). Data are mean ± s.d. (n = 5 plants). Numbers indicate P values derived from a two-tailed t-test when compared with VC. GA treatment was performed on the plants 1 week after germination for 3 weeks. e, Gross morphology of ACE1-RNAi in B. distachyon without (left) and with (right) GA treatment. Numbers below the photographs represent independent T0 lines. Scale bars, 5 cm. f, Sequence alignment of ACE1 homologues of B. distachyon. Red arrows indicate the positions of primers for detection of the expression of endogenous ACE1 homologues. g, Expression of ACE1 homologues in B. distachyon in VC and ACE1-RNAi lines (n = 4 plants). h, i, Total internode length without (h, n = 9 plants) and with (i, n = 10 plants) GA treatment. Data are mean ± s.d. and numbers indicate P values derived from a two-tailed t-test when compared with VC (g–i). One week after seed germination, plants were treated with GA for 3 weeks. j, Gross morphology of vector control and ACE1C9285ox in barley in the mock condition. Right panels show enlarged views of internodes. Arrowheads indicate the positions of nodes. k, Expression of ACE1 in VC and of ACE1C9285ox in barley (n = 4 plants). Number indicates P value derived from a two-tailed t-test. l, m, Total internode length (l) and number of elongated internodes (m) in transgenic barley plants overexpressing ACE1C9285ox. Data are mean ± s.d. (n = 6 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. There were significant differences in total internode length and number of elongated internodes. Plastochrons were not affected by ACE1C9285ox, suggesting that initiation of internode elongation is promoted in plants overexpressing ACE1C9285ox. n, Schematic of plant growth of sugarcane and GA treatment. After excising the leaves of the T0 plant, the elongated internode was cut into 3 cm pieces, which included a node (left). Cut internodes were divided into two and planted in soil. Plants with two or three developed leaves were subjected to mock or GA treatment for 4 weeks (right). o, Expression of ACE1 in VC and of ACE1C9285ox in sugarcane. Data are mean ± s.d. (n = 4 plants). Number indicates P value derived from a two-tailed t-test. p, Total internode length and number of elongated internodes in transgenic sugarcane plants overexpressing ACE1C9285ox. Data are mean ± s.d. Numbers indicate P values derived from a two-tailed t-test in -GA or +GA. The number of elongated internodes differed significantly between the control and ACE1C9285ox after mock and GA treatments. q, Sequence alignment of ACE1 homologues in sugarcane. Sequences were obtained from Sugarcane Genome Hub (https://sugarcane-genome.cirad.fr). All experiments were repeated twice with similar results.
Extended Data Fig. 11 The identification of the causal gene in the QTL on chromosome 12 in response to GA.
a, Total internode length of plants that overexpress SK1 or SK2 after maturation. Dots indicate the total internode lengths of individual plants. Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. b, Genotype of the candidate region on chromosome 12. The mapping population generated by crossing NIL3+12 and NIL3 possessed a C9285 homozygote on chromosome 3. We previously identified roles for SK1 and SK2 on chromosome 12 in the deep-water response; however, positional cloning in response to GA excluded SNORKEL genes. Red arrow, candidate region of the GA response. T65-Homo, Hetero and C9285-Homo indicate T65-type homozygous, heterozygous and C9285-type homozygous genotypes, respectively, of the candidate region. c, Total internode length of each genotype after 3 weeks of GA treatment from germination. Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. d, Length of each internode. Ordinal number, internode position after germination. Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. e, f, Expression levels of candidate genes before (0 h) or after 24 h of 10−4 M GA treatment. Data are mean ± s.d. (n = 3 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. g, Gross morphology of vector control (VC) and LOC_Os12g42250 (DEC1T65)-ox in T65 genetic background after the heading stage. Right panels show enlarged views of internodes and basal region of LOC_Os12g42250 (DEC1T65)-ox (dashed square). The number of non-elongated internodes increased, and internode length decreased, in LOC_Os12g42250 (DEC1T65)-ox, resulting in a reduction in total internode length. h, Effect of LOC_Os12g42250 (DEC1T65)-ox expression in T65 genetic background on total internode length. Y-axis, expression level of LOC_Os12g42250 (DEC1T65)-ox; x-axis, total internode length. Dots indicate individual plants (n = 5 plants). Right, an enlarged view of the dashed square in the left panel. Plants with high LOC_Os12g42250 (DEC1T65)-ox expression showed a tendency to have a shorter plant height. i, Gross morphology of plants overexpressing LOC_Os12g42260 in the NIL12 background (T0). Arrowheads indicate the positions of nodes. One-month-old plants were treated with GA for 2 weeks. Scale bars, 10 cm. Right, internode length of the plants that overexpress LOC_Os12g42260 in the NIL12 genetic background (T0). Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. j, Coding sequences of T65 DEC1 and C9285 DEC1. k, Amino acid sequences of T65 DEC1 and C9285 DEC1. Green and red lines, LxLxL-type EAR motif and C2H2 zinc-finger domain, respectively. l, Gross morphology of the control plant and LOC_Os12g42250 (dec1) mutant generated by CRISPR/Cas9 in NIL12 background (T0) in the absence of GA. Arrowheads indicate the positions of nodes. Scale bars, 10 cm. m, Expression level of DEC1, OsMADS14 and OsMADS15 at each growth stage of Nipponbare. Data are mean ± s.d. (n = 4 plants). Numbers indicate P values derived from a two-tailed t-test when compared with the 4-LS. OsMADS14 and OsMADS15 were expressed at the 8-LS, whereas DEC1 expression began to decrease at the 8-LS in Nipponbare. All experiments were repeated twice with similar results.
Extended Data Fig. 12 Expression level, subcellular localization and transcriptional activity of DEC1.
a, Schematic diagram of elongating internode of C9285 in shallow-water (left) and deep-water (right) conditions. Deep-water treatment was conducted for 24 h. Numbers next to internodes indicate the sampling positions and corresponding distances from point 1 (0–0.5 cm), the node at the base of the axillary bud; 2, 0.5–1.0 cm; 3, 1.0–1.5 cm; 4, 1.5–3.0 cm; 5, 3.0–4.0 cm from position 1. Point 6 is 4.5–6.0 cm in shallow-water conditions, and 7.5–9.0 cm in deep-water conditions from position 1, respectively. Right panels, expression of DEC1 and the cell-division-related genes, HISTONE H4 and CDKA1. DEC1 was highly expressed in the cell-division zone at around positions 2 and 3 in shallow-water conditions but was downregulated in deep-water conditions. By contrast, the expression of cell-division-related genes was increased at positions 2 and 3 in deep-water conditions. Data are mean ± s.d. (n = 3 plants). b, Phylogenetic tree of DEC1 homologues constructed using the SALAD database (https://salad.dna.affrc.go.jp/salad/en/). c, Diagram of conserved motifs constructed using the SALAD database and the DEC1 sequence. Boxes and numbers represent conserved motifs. d, Conserved sequence of motif number 8 in c. This motif contains an LxLxL-type EAR motif. e, Conserved sequence of motif number 1 in c. This motif contains a C2H2-type zinc-finger domain. f, Conserved sequence of motif number 8 in c. This motif contains an LxLxL-type EAR motif. Conserved motifs were detected by WebLogo (http://weblogo.berkeley.edu). g, Subcellular localization of GFP, DEC1T65–GFP and DEC1C9285–GFP in onion epidermal cells. The signal of 35S:mCherry was used as a control. In the fluorescence profile, the GFP signal of 35S:GFP overlapped with the mCherry signal, whereas 35S:DEC1T65-GFP and 35S:DEC1C9285-GFP showed a high intensity of GFP signal in the nuclei, suggesting that DEC1T65 and DEC1C9285 are localized in the nuclei. Fluorescence profiles were imaged by the profile tool of the ZEN software (Carl Zeiss). Plasmid constructs were introduced by particle bombardment. h, Transcriptional activity of DEC1 by yeast one-hybrid assay. Left, diagrams of effector constructs. All experiments were repeated twice with similar results.
Extended Data Fig. 13 The effect of DEC1 on internode elongation.
a, Gross morphology of vector control plants and plants that overexpress DEC1T65 (DEC1T65ox) or DEC1C9285 (DEC1C9285ox) in barley. Right panels show an enlarged view of the basal region of DEC1T65ox and DEC1C9285ox. Arrow indicates the position of the shoot apex. b–d, DEC1 expression (b), plant height (c) and total internode length (d) of transgenic barley plants overexpressing DEC1. Data are mean ± s.d. (n = 4 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. e, Internode length of dec1 mutant (T1). Internodes of 5-leaf-stage plants of VC and dec1 mutant were measured after 3 and 10 days of 10−4 M GA treatment. Data are mean ± s.d. (n = 6 plants). Numbers indicate P values derived from a two-tailed t-test when compared with total internode length of mock control. f, g, EdU signal in the dec1 mutant after mock or GA treatment for 3 days (f) and 10 days (g). The fifth internodes of 5-leaf stage plants are highlighted (f). An EdU signal was detected at the basal region of elongating internodes after 10 days of GA treatment (g). h, Expression of ACE1 and DEC1 in ACE1C9285ox and the dec1 mutant in T65 genetic background. Data are mean ± s.d. (n = 4 plants). One-way ANOVA followed by Tukey’s multiple-comparison test. i, j, Effect of overexpression of SNORKEL genes on ACE1 and DEC1 expression in NIL1+3+12. To examine the effect of SNORKEL genes on ACE1 and DEC1 expression, SNORKEL genes were overexpressed in NIL1+3+12 possessing C9285-type ACE1 and DEC1. SKox plants were grown in shallow-water conditions. i, Expression of SK1, SK2, ACE1 and DEC1 in SKox/NIL1+3+12 and C9285 in shallow-water and deep-water conditions Data are mean ± s.d. (n = 3 plants). j, Total internode length of SKox plants in shallow-water conditions. Data are mean ± s.d. (n = 3 plants). Numbers indicate P values derived from a two-tailed t-test when compared with VC (i, j). All experiments were repeated twice with similar results.
Extended Data Fig. 14 The selection of ACE1 and DEC1, and application for breeding.
a, Multiple regression of ACE1 and DEC1 in 28 individuals of O. rufipogon by generalized linear model (GLM) regression analysis. The two-sided P value of a test on the null hypothesis of that the coefficient is equal to zero (no effect), was calculated with F-test which describes the ratio of two chi-squared distributed variables. b, Transition of EHH from the C9285-like (red) and T65-like (blue) haplotypes of DEC1, in 28 O. rufipogon. The two-sided P value of a test on the null hypothesis of no selection from each core SNP was calculated from the Z-scores of the iHS values in chromosome 12 using the cumulative probability density function of the normal distribution. c, d, Comparison of allele frequencies of DEC1 (c) and ACE1 (d), in O. rufipogon and six subpopulations of O. sativa. e, Genotypes of T65 and NIL1+3+12. NIL1+3+12 possesses QTL regions on chromosomes 1, 3 and 12 for the deep-water response in T65 genetic background. f, Gross morphology of T65 and NIL1+3+12. Two-month old plants underwent shallow-water or deep-water treatment. During deep-water treatment, the water level was increased by 10 cm per day until a depth of 120 cm was achieved and continued until seed maturation (about 3 months). The water depth of conditions was 5 cm from the soil surface. g, h, Internode length after ripening (g) and number of grains per panicle (h). Data are mean ± s.d. The number of plants (n) is shown. One-way ANOVA followed by Tukey’s multiple-comparison test. T65 died in deep-water conditions within 3 months and was non-quantifiable (g, h). All experiments were repeated twice with similar results. i, Schematic of the antagonistic regulatory mechanism of internode elongation by ACE1/ACL1 and DEC1. Top and bottom panels are models for normal paddy rice and deepwater rice, respectively.
Supplementary information
Supplementary Information
This file provides information, results and discussions omitted in the main text. This file contains 10 sections that are cited at appropriate points in the main text, associated references, and Supplementary Figure 1 (raw gel images).
Supplementary Table
Supplementary Table 1. The list of probes detected by trans-eQTL analysis related to the ACE1 expression.
Supplementary Table
Supplementary Table 2. Sequence alignment of the motifs in ACE1C9285 and the homologs.
Supplementary Table
Supplementary Table 3. Genotype and internode length in deepwater of O. rufipogon. 0, T65 type allele; 1, C9285 allele.
Supplementary Table
Supplementary Table 4. Genotype of ACE1 and DEC1 in O. sativa population. 0, Nipponbare type allele; 1, C9285 allele.
Supplementary Table
Supplementary Table 5. Primer list.
Video 1
Time-lapse movie of C9285 internode elongation in response to GA. A section of a C9285 (DW rice) plant containing internodes and the basal region of the leaf sheath was treated with 0.5 x 10−4 M GA3 for 7 days. Arrowheads indicate the positions of nodes. The experiments were repeated three times with similar results.
Source data
Rights and permissions
About this article
Cite this article
Nagai, K., Mori, Y., Ishikawa, S. et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 584, 109–114 (2020). https://doi.org/10.1038/s41586-020-2501-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2501-8
This article is cited by
-
Strong culm: a crucial trait for developing next-generation climate-resilient rice lines
Physiology and Molecular Biology of Plants (2024)
-
OsNAC103, an NAC transcription factor negatively regulates plant height in rice
Planta (2024)
-
Oxygenation alleviates waterlogging-caused damages to cherry rootstocks
Molecular Horticulture (2023)
-
Identification of Genomic Regions for Deep-Water Resistance in Rice for Efficient Weed Control with Reduced Herbicide Use
Rice (2023)
-
Can the Wild Perennial, Rhizomatous Rice Species Oryza longistaminata be a Candidate for De Novo Domestication?
Rice (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.