Abstract
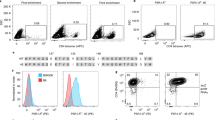
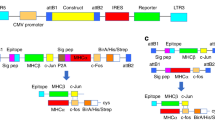
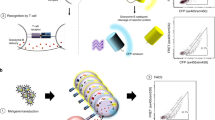
Peptide–major histocompatibility complex (pMHC) multimers enable the detection of antigen-specific T cells in studies ranging from vaccine efficacy to cancer immunotherapy. However, this technology is unreliable when applied to pMHC class II for the detection of CD4+ T cells. Here, using a combination of molecular biological and immunological techniques, we cloned sequences encoding human leukocyte antigen (HLA)-DP, HLA-DQ and HLA-DR molecules with enhanced CD4 binding affinity (with a Kd of 8.9 ± 1.1 µM between CD4 and affinity-matured HLA-DP4) and produced affinity-matured class II dimers that stain antigen-specific T cells better than conventional multimers in both in vitro and ex vivo analyses. Using a comprehensive library of dimers for HLA-DP4, which is the most frequent HLA allele in many ancestry groups, we mapped 103 HLA-DP4-restricted epitopes derived from diverse tumor-associated antigens and cloned the cognate T-cell antigen receptor (TCR) genes from in vitro-stimulated CD4+ T cells. The availability of affinity-matured class II dimers across HLA-DP, HLA-DQ and HLA-DR alleles will aid in the investigation of human CD4+ T-cell responses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The main data of this study are available within the article and its Supplementary Figures. Source data are provided with this paper. All other data are available from the corresponding author upon reasonable request.
References
Seder, R. A. & Ahmed, R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 4, 835–842 (2003).
Jonsson, P. et al. Remarkably low affinity of CD4/peptide–major histocompatibility complex class II protein interactions. Proc. Natl Acad. Sci. USA 113, 5682–5687 (2016).
Xiong, Y., Kern, P., Chang, H. & Reinherz, E. T cell receptor binding to a pMHCII ligand is kinetically distinct from and independent of CD4. J. Biol. Chem. 276, 5659–5667 (2001).
Davis, S. J. et al. The nature of molecular recognition by T cells. Nat. Immunol. 4, 217–224 (2003).
Garcia, K. C. et al. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature 384, 577–581 (1996).
Wyer, J. R. et al. T cell receptor and coreceptor CD8α bind peptide–MHC independently and with distinct kinetics. Immunity 10, 219–225 (1999).
Altman, J. D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
Meyer, A. L. et al. Direct enumeration of Borrelia-reactive CD4 T cells ex vivo by using MHC class II tetramers. Proc. Natl Acad. Sci. USA 97, 11433–11438 (2000).
Vollers, S. S. & Stern, L. J. Class II major histocompatibility complex tetramer staining: progress, problems, and prospects. Immunology 123, 305–313 (2008).
Nepom, G. T. MHC class II tetramers. J. Immunol. 188, 2477–2482 (2012).
Wooldridge, L. et al. Tricks with tetramers: how to get the most from multimeric peptide–MHC. Immunology 126, 147–164 (2009).
Zhao, W. & Sher, X. Systematically benchmarking peptide–MHC binding predictors: from synthetic to naturally processed epitopes. PLoS Comput. Biol. 14, e1006457 (2018).
Wang, X. X. et al. Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4–HLA-DR1 complex. Proc. Natl Acad. Sci. USA 108, 15960–15965 (2011).
Konig, R., Huang, L. Y. & Germain, R. N. MHC class II interaction with CD4 mediated by a region analogous to the MHC class I binding site for CD8. Nature 356, 796–798 (1992).
Konig, R. Interactions between MHC molecules and co-receptors of the TCR. Curr. Opin. Immunol. 14, 75–83 (2002).
Artyomov, M. N., Lis, M., Devadas, S., Davis, M. M. & Chakraborty, A. K. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc. Natl Acad. Sci. USA 107, 16916–16921 (2010).
Solberg, O. D. et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 69, 443–464 (2008).
Yamashita, Y. et al. HLA-DP84Gly constitutively presents endogenous peptides generated by the class I antigen processing pathway. Nat. Commun. 8, 15244 (2017).
Anczurowski, M. et al. Mechanisms underlying the lack of endogenous processing and CLIP-mediated binding of the invariant chain by HLA-DP84Gly. Sci. Rep. 8, 4804 (2018).
Pinto, E. M. et al. Prognostic significance of major histocompatibility complex class II expression in pediatric adrenocortical tumors: a St. Jude and Children’s Oncology Group study. Clin. Cancer Res. 22, 6247–6255 (2016).
Yao, X. et al. Isolation and characterization of an HLA-DPB1*04:01-restricted MAGE-A3 T-cell receptor for cancer immunotherapy. J. Immunother. 39, 191–201 (2016).
Lu, Y. C. et al. Treatment of patients with metastatic cancer using a major histocompatibility complex class II-restricted T-cell receptor targeting the cancer germline antigen MAGE-A3. J. Clin. Oncol. 35, 3322–3329 (2017).
Merkel, P. A. et al. Identification of functional and expression polymorphisms associated with risk for antineutrophil cytoplasmic autoantibody-associated vasculitis. Arthritis Rheumatol. 69, 1054–1066 (2017).
Lamprecht, P. et al. Pathogenetic and clinical aspects of anti-neutrophil cytoplasmic autoantibody-associated vasculitides. Front. Immunol. 9, 680 (2018).
Hilhorst, M. et al. HLA-DPB1 as a risk factor for relapse in antineutrophil cytoplasmic antibody-associated vasculitis: a cohort study. Arthritis Rheumatol. 68, 1721–1730 (2016).
Williams, T. M. Human leukocyte antigen gene polymorphism and the histocompatibility laboratory. J. Mol. Diagn. 3, 98–104 (2001).
Melenhorst, J. J. et al. Detection of low avidity CD8+ T cell populations with coreceptor-enhanced peptide–major histocompatibility complex class I tetramers. J. Immunol. Methods 338, 31–39 (2008).
Wooldridge, L. et al. Interaction between the CD8 coreceptor and major histocompatibility complex class I stabilizes T cell receptor–antigen complexes at the cell surface. J. Biol. Chem. 280, 27491–27501 (2005).
Wooldridge, L. et al. MHC class I molecules with superenhanced CD8 binding properties bypass the requirement for cognate TCR recognition and nonspecifically activate CTLs. J. Immunol. 184, 3357–3366 (2010).
Butler, M. O. et al. Ex vivo expansion of human CD8+ T cells using autologous CD4+ T cell help. PLoS ONE 7, e30229 (2012).
Heemskerk, M. H. et al. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood 102, 3530–3540 (2003).
Straetemans, T. et al. TCR gene transfer: MAGE-C2/HLA-A2 and MAGE-A3/HLA-DP4 epitopes as melanoma-specific immune targets. Clin. Dev. Immunol. 2012, 586314 (2012).
Lin, Y. et al. HLA-DPB1*05:01-restricted WT1332-specific TCR-transduced CD4+ T lymphocytes display a helper activity for WT1-specific CTL induction and a cytotoxicity against leukemia cells. J. Immunother. 36, 159–170 (2013).
Odunsi, K. et al. Vaccination with an NY-ESO-1 peptide of HLA class I/II specificities induces integrated humoral and T cell responses in ovarian cancer. Proc. Natl Acad. Sci. USA 104, 12837–12842 (2007).
Laurin, D. et al. Minor histocompatibility antigen DDX3Y induces HLA-DQ5-restricted T cell responses with limited TCR-Vβ usage both in vivo and in vitro. Biol. Blood Marrow Transplant 12, 1114–1124 (2006).
Jones, C. M., Lake, R. A., Lamb, J. R. & Faith, A. Degeneracy of T cell receptor recognition of an influenza virus hemagglutinin epitope restricted by HLA-DQ and -DR class II molecules. Eur. J. Immunol. 24, 1137–1142 (1994).
Hennecke, J., Carfi, A. & Wiley, D. C. Structure of a covalently stabilized complex of a human αβ T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J. 19, 5611–5624 (2000).
Tsuji, T. et al. Heat shock protein 90-mediated peptide-selective presentation of cytosolic tumor antigen for direct recognition of tumors by CD4+ T cells. J. Immunol. 188, 3851–3858 (2012).
Neunkirchner, A. et al. Human TCR transgenic Bet v 1-specific Th1 cells suppress the effector function of Bet v 1-specific Th2 cells. J. Immunol. 187, 4077–4087 (2011).
Benati, D. et al. Public T cell receptors confer high-avidity CD4 responses to HIV controllers. J. Clin. Invest. 126, 2093–2108 (2016).
Nakatsugawa, M. et al. CD4+ and CD8+ TCRβ repertoires possess different potentials to generate extraordinarily high-avidity T cells. Sci. Rep. 6, 23821 (2016).
Nakatsugawa, M. et al. Specific roles of each TCR hemichain in generating functional chain-centric TCR. J. Immunol. 194, 3487–3500 (2015).
Ochi, T. et al. Optimization of T-cell reactivity by exploiting TCR chain centricity for the purpose of safe and effective antitumor TCR gene therapy. Cancer Immunol. Res. 3, 1070–1081 (2015).
Hirano, N. et al. Engagement of CD83 ligand induces prolonged expansion of CD8+ T cells and preferential enrichment for antigen specificity. Blood 107, 1528–1536 (2006).
Butler, M. O. et al. Long-lived antitumor CD8+ lymphocytes for adoptive therapy generated using an artificial antigen-presenting cell. Clin. Cancer Res. 13, 1857–1867 (2007).
Hirano, N. et al. Efficient presentation of naturally processed HLA class I peptides by artificial antigen-presenting cells for the generation of effective antitumor responses. Clin. Cancer Res. 12, 2967–2975 (2006).
Wooldridge, L. et al. Anti-coreceptor antibodies profoundly affect staining with peptide–MHC class I and class II tetramers. Eur. J. Immunol. 36, 1847–1855 (2006).
Hirano, N. et al. Identification of an immunogenic CD8+ T-cell epitope derived from γ-globin, a putative tumor-associated antigen for juvenile myelomonocytic leukemia. Blood 108, 2662–2668 (2006).
Imataki, O. et al. IL-21 can supplement suboptimal Lck-independent MAPK activation in a STAT-3-dependent manner in human CD8+ T cells. J. Immunol. 188, 1609–1619 (2012).
Su, L. F., Kidd, B. A., Han, A., Kotzin, J. J. & Davis, M. M. Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity 38, 373–383 (2013).
Bienert, S. et al. The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res. 45, D313–D319 (2017).
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L. & Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7, 10480 (2017).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Acknowledgements
This work was supported by the Ontario Institute for Cancer Research Clinical Investigator Award IA-039 (N.H.), the Princess Margaret Cancer Centre Innovation Accelerator Fund (N.H.), the Ira Schneider Memorial Cancer Research Foundation (N.H.), the Princess Margaret Cancer Foundation (N.H. and M.O.B.), the Uehara Memorial Foundation Research Fellowship Program (K. Sugata), the Mitacs Internship (K.M.), the Japan Society for the Promotion of Science Postdoctoral Fellowship for Overseas Researchers and the Guglietti fellowship (Y.K.), the Province of Ontario (T.G. and M.A.), the Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship (T.G.) and the Frederick Banting and Charles Best Canada Graduate Scholarship (C.-H.W.).
Author information
Authors and Affiliations
Contributions
K. Sugata, Y.M., Y.Y., M.N. and N.H. designed the project. K. Sugata, Y.M., Y.Y., M.N., T.G., L.H., K. Saso, M.A.R., M.A., C.-H.W., K.M., H.S., Y.K., Y.O., D.L. and B.D.B. performed the experiments. M.O.B. provided critical human samples. T.W.M. provided critical resources. N.H. administered and supervised the project. K. Sugata and N.H. analyzed the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.O.B. has served on advisory boards for Merck, BMS, Novartis, GSK, Immunocore, Immunovaccine, Sanofi and EMD Serono and received research funding for investigator-initiated clinical trials from Merck and Takara Bio. N.H. has received research funding from Takara Bio and served as a consultant for Takara Bio. The University Health Network has filed a patent application related to this study on which N.H., K. Sugata, Y.Y., M.N., K. Saso, M.A.R. and T.G. are named as inventors. T.W.M. and N.H. are cofounders and have equity in TCRyption to which the technologies used in this study have been licensed.
Additional information
Peer review information Nature Biotechnology thanks Kari C. Nadeau and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–38.
Supplementary Data
Supplementary Tables 1–3, statistical source data for the Supplementary Figures and structural models for Supplementary Figs. 3 and 31.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Rights and permissions
About this article
Cite this article
Sugata, K., Matsunaga, Y., Yamashita, Y. et al. Affinity-matured HLA class II dimers for robust staining of antigen-specific CD4+ T cells. Nat Biotechnol 39, 958–967 (2021). https://doi.org/10.1038/s41587-021-00836-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-00836-4
This article is cited by
-
Nanoparticle-based modulation of CD4+ T cell effector and helper functions enhances adoptive immunotherapy
Nature Communications (2022)
-
Recent advances in cancer immunotherapy
Discover Oncology (2021)