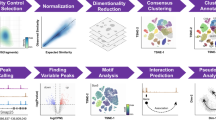
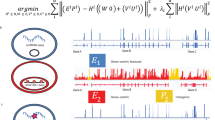
Abstract
Integrating large single-cell gene expression, chromatin accessibility and DNA methylation datasets requires general and scalable computational approaches. Here we describe online integrative non-negative matrix factorization (iNMF), an algorithm for integrating large, diverse and continually arriving single-cell datasets. Our approach scales to arbitrarily large numbers of cells using fixed memory, iteratively incorporates new datasets as they are generated and allows many users to simultaneously analyze a single copy of a large dataset by streaming it over the internet. Iterative data addition can also be used to map new data to a reference dataset. Comparisons with previous methods indicate that the improvements in efficiency do not sacrifice dataset alignment and cluster preservation performance. We demonstrate the effectiveness of online iNMF by integrating more than 1 million cells on a standard laptop, integrating large single-cell RNA sequencing and spatial transcriptomic datasets, and iteratively constructing a single-cell multi-omic atlas of the mouse motor cortex.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
• Human PBMC from Kang et al.9 (GSE96583) distributed by SeuratData
• Human pancreatic islet cells from Grün et al.10 (GSE81076), Muraro et al.11 (GSE85241), Lawlor et al.12 (GSE86469), Baron et al.13 (GSE84133) and Segerstolpe et al.14 (E-MTAB-5061) distributed by SeuratData
• Adult mouse brain cells from Saunders et al.7 (http://dropviz.org/)
• Mouse Organogenesis Cell Atlas from Cao et al.18 (https://oncoscape.v3.sttrcancer.org/atlas.gs.washington.edu.mouse.rna/downloads)
• Mouse hippocampus cells from Rodriques et al.19 (https://singlecell.broadinstitute.org/single_cell/study/SCP354/slide-seq-study#study-download)
• Mouse hippocampus cells from Yao et al.22 (http://data.nemoarchive.org/biccn/grant/zeng/zeng/transcriptome/scell/10X/processed/YaoHippo2020/)
• Mouse hypothalamic pre-optic region data from Moffitt et al.23 (https://datadryad.org/stash/dataset/doi:10.5061/dryad.8t8s248 and GSE113576)
• Mouse primary motor cortex cells from Yao et al.27 (https://assets.nemoarchive.org/dat-ch1nqb7)
Code availability
An R implementation of LIGER is available from the Comprehensive R Archive Network at https://cran.r-project.org/package=rliger and on GitHub at https://github.com/welch-lab/liger, along with detailed installation instructions. Tutorials demonstrating package functionality, including online learning for Scenario 1, Scenario 2 and Scenario 3, are available on the GitHub page.
References
Ye, Z. & Sarkar, C. A. Towards a quantitative understanding of cell identity. Trends Cell Biol. 28, 1030–1048 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Welch, J. D. et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887 (2019).
Mairal, J., Bach, F., Ponce, J. & Sapiro, G. Online learning for matrix factorization and sparse coding. J. Mach. Learn. Res. 11, 19–60 (2010).
Saunders, A. et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030 (2018).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94 (2018).
Grün, D. et al. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19, 266–277 (2016).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394 (2016).
Lawlor, N. et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 27, 208–222 (2017).
Baron, M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360 (2016).
Segerstolpe, Å. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016).
Toda, T., Parylak, S. L., Linker, S. B. & Gage, F. H. The role of adult hippocampal neurogenesis in brain health and disease. Mol. Psychiatry 24, 67–87 (2019).
Ernst, A. et al. Neurogenesis in the striatum of the adult human brain. Cell 156, 1072–1083 (2014).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014 (2018).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Preprint at bioRxiv https://doi.org/10.1101/2020.03.30.015214 (2020).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018).
Ecker, J. R. et al. The BRAIN Initiative Cell Census Consortium: lessons learned toward generating a comprehensive brain cell atlas. Neuron 96, 542–557 (2017).
HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
Yao, Z. et al. An integrated transcriptomic and epigenomic atlas of mouse primary motor cortex cell types. Preprint at bioRxiv https://doi.org/10.1101/2020.02.29.970558 (2020).
Tasic, B. et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346 (2016).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Büttner, M., Miao, Z., Wolf, F. A., Teichmann, S. A. & Theis, F. J. A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods 16, 43–49 (2019).
Hubert, L. & Arabie, P. Comparing partitions. J. Classification 2, 193–218 (1985).
Rand, W. M. Objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc. 66, 846–850 (1971).
Zappia, L., Phipson, B. & Oshlack, A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 18, 174 (2017).
Acknowledgements
This work was supported by National Institutes of Health grants R01AI149669-01, R01HG010883-01 and RF1MH123199 (to J.D.W.) and 5U19MH114831 (to J.R.E.). J.R.E. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
S.P., C.L., R.C., J.S., A.R., J.R.N., M.M.B., J.R.E. and B.R. generated the snATAC-seq and snmC-seq data. J.D.W. conceived the idea of online iNMF. C.G. and J.D.W. developed and implemented the online iNMF algorithm. C.G., J.L., A.R.K. and J.D.W. carried out data analyses. C.G., J.L., A.R.K. and J.D.W. wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
A patent application on LIGER has been submitted by the Broad Institute and the General Hospital Corporation with J.D.W. listed as an inventor. The remaining authors declare no competing financial interests.
Additional information
Peer review information Nature Biotechnology thanks Samantha A. Morris and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Convergence behavior for online iNMF and batch iNMF algorithms on scRNA-seq data from the adult mouse brain, human PBMC and human pancreas.
The online iNMF algorithm exhibits faster convergence and better objective minimization after a fixed amount of training time. The advantage of the online algorithm in convergence speed is more apparent for larger datasets. a-c, Adult mouse brain (n = 691,962 cells, nine individual datasets). d-f, Human PBMC (n = 13,999 cells, two individual datasets). g-i, Human pancreas (n = 14,890 cells, eight individual datasets). Center lines of box plots show the median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; and points are outliers.
Extended Data Fig. 2 Online and batch iNMF yield highly similar UMAP visualizations.
We performed online iNMF and batch iNMF on data from mouse cortex (n = 255,353 cells), human PBMC (n = 13,999 cells), and human pancreas (n = 14,890 cells). Online iNMF and batch iNMF produce very similar visualizations, suggesting that the approaches give very similar dataset alignment and cluster preservation. We subsequently confirmed this qualitative observation using quantitative metrics.
Extended Data Fig. 3
Benchmarking integration across data modalities (RNA+ATAC). 5,000 cells from the snRNA-seq dataset and 5,000 cells from the snATAC-seq dataset from MOp data collection were integrated using four different methods. The cells are exhibited in 2-dimensional UMAP space and colored by dataset.
Extended Data Fig. 4 Performing online iNMF in three scenarios produces similar results.
These analyses were carried out separately to integrate eight MOp datasets (scRNA-seq, snRNA-seq, snATAC-seq and snmC-seq, n = 408,885) using online iNMF in scenario 1 (a), scenario 2 (b), and scenario 3 (c). The results are visualized in UMAP coordinates and the cells are colored by the cell type annotations from Fig. 6.
Supplementary information
Supplementary Information
Supplementary Notes, Figs. 1–15 and Table 1
Rights and permissions
About this article
Cite this article
Gao, C., Liu, J., Kriebel, A.R. et al. Iterative single-cell multi-omic integration using online learning. Nat Biotechnol 39, 1000–1007 (2021). https://doi.org/10.1038/s41587-021-00867-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-00867-x
This article is cited by
-
Semi-supervised integration of single-cell transcriptomics data
Nature Communications (2024)
-
Online mode development of Korean art learning in the post-epidemic era based on artificial intelligence and deep learning
The Journal of Supercomputing (2024)
-
Spatial omics technologies at multimodal and single cell/subcellular level
Genome Biology (2022)
-
Multi-omics single-cell data integration and regulatory inference with graph-linked embedding
Nature Biotechnology (2022)
-
Spatial components of molecular tissue biology
Nature Biotechnology (2022)