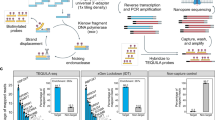
Abstract
Current next-generation RNA-sequencing (RNA-seq) methods do not provide accurate quantification of small RNAs within a sample, due to sequence-dependent biases in capture, ligation and amplification during library preparation. We present a method, absolute quantification RNA-sequencing (AQRNA-seq), that minimizes biases and provides a direct, linear correlation between sequencing read count and copy number for all small RNAs in a sample. Library preparation and data processing were optimized and validated using a 963-member microRNA reference library, oligonucleotide standards of varying length, and RNA blots. Application of AQRNA-seq to a panel of human cancer cells revealed >800 detectable miRNAs that varied during cancer progression, while application to bacterial transfer RNA pools, with the challenges of secondary structure and abundant modifications, revealed 80-fold variation in tRNA isoacceptor levels, stress-induced site-specific tRNA fragmentation, quantitative modification maps, and evidence for stress-induced, tRNA-driven, codon-biased translation. AQRNA-seq thus provides a versatile means to quantitatively map the small RNA landscape in cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All sequencing and proteomics data that support the findings of this study have been variously deposited in public databases: RNA-seq studies reported in Figs. 3–5, Supplementary Figs. 2, 4 and 8 and the proteomics studies reported in Fig. 4 and Supplementary Fig. 6 have been deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA579244; miRNA and standards data shown in Fig. 2 have been deposited in the Gene Expression Omnibus (GEO) under accession no. GSE139936; and data for miRNA studies in HMEC cells shown in Fig. 6 have been deposited in GEO as accession no. GSE159434.
Code availability
The software used in the studies presented here is publicly available as follows. Blast v.2.6.0 (nucleotide BLAST) available at https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastDocs&DOC_TYPE=Download. Peakfit.m v.9.0 available at Tom O’Haver, MATLAB Central File Exchange: https://terpconnect.umd.edu/~toh/spectrum/. fgrep (Linux command) available at https://unix.stackexchange.com/questions/17949/what-is-the-difference-between-grep-egrep-and-fgrep. fastxtoolkit v.0.013 available at http://hannonlab.cshl.edu/fastx_toolkit/. Custom python scripts are available at GitHub https://github.com/dedonlab/ (https://github.com/dedonlab/aqrnaseq for prokaryotic process scripts and https://github.com/dedonlab/general_aqrnaseq for eukaryotic/general pipeline scripts).
References
Wang, Z., Gerstein, M. & Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009).
Cech, T. R. & Steitz, J. A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94 (2014).
Hafner, M. et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 17, 1697–1712 (2011).
Zhang, Z., Lee, J. E., Riemondy, K., Anderson, E. M. & Yi, R. High-efficiency RNA cloning enables accurate quantification of miRNA expression by deep sequencing. Genome Biol. 14, R109 (2013).
Fuchs, R. T., Sun, Z., Zhuang, F. & Robb, G. B. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS ONE 10, e0126049 (2015).
Alon, S. et al. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 21, 1506–1511 (2011).
Zhuang, F., Fuchs, R. T., Sun, Z., Zheng, Y. & Robb, G. B. Structural bias in T4 RNA ligase-mediated 3’-adapter ligation. Nucleic Acids Res. 40, e54 (2012).
Pang, Y. L., Abo, R., Levine, S. S. & Dedon, P. C. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 42, e170 (2014).
Linsen, S. E. et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods 6, 474–476 (2009).
Machnicka, M. A., Olchowik, A., Grosjean, H. & Bujnicki, J. M. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 11, 1619–1629 (2014).
Björk, G. R. et al. Transfer RNA modification. Annu. Rev. Biochem. 56, 263–287 (1987).
Motorin, Y. & Helm, M. Methods for RNA modification mapping using deep sequencing: established and new emerging technologies. Genes (Basel) 10, 35 (2019).
Motorin, Y., Muller, S., Behm‐Ansmant, I. & Branlant, C. Identification of modified residues in RNAs by reverse transcription‐based methods. Methods Enzymol. 425, 21–53 (2007).
Shigematsu, M. et al. YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. Nucleic Acids Res. 45, e70 (2017).
Gogakos, T. et al. Characterizing expression and processing of precursor and mature human tRNAs by Hydro-tRNAseq and PAR-CLIP. Cell Rep. 20, 1463–1475 (2017).
Kim, H. et al. Bias-minimized quantification of microRNA reveals widespread alternative processing and 3’ end modification. Nucleic Acids Res. 47, 2630–2640 (2019).
Dai, Q., Zheng, G., Schwartz, M. H., Clark, W. C. & Pan, T. Selective enzymatic demethylation of N(2),N(2) -dimethylguanosine in RNA and its application in high-throughput tRNA sequencing. Angew. Chem. Int. Ed. Engl. 56, 5017–5020 (2017).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Xu, H., Yao, J., Wu, D. C. & Lambowitz, A. M. Improved TGIRT-seq methods for comprehensive transcriptome profiling with decreased adapter dimer formation and bias correction. Sci. Rep. 9, 7953 (2019).
Mohr, S. et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 19, 958–970 (2013).
Jayaprakash, A. D., Jabado, O., Brown, B. D. & Sachidanandam, R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 39, e141 (2011).
Lovett, S. T. & Kolodner, R. D. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl Acad. Sci. USA 86, 2627–2631 (1989).
Shepherd, J. & Ibba, M. Bacterial transfer RNAs. FEMS Microbiol. Rev. 39, 280–300 (2015).
Ardell, D. H. & Hou, Y. M. Initiator tRNA genes template the 3’ CCA end at high frequencies in bacteria. BMC Genomics 17, 1003 (2016).
Kozomara, A. & Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014).
Herbert, Z. T. et al. Multisite evaluation of next-generation methods for small RNA quantification. J. Biomol. Tech. 31, 47–56 (2020).
Coenen-Stass, A. M. L. et al. Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol. 15, 1133–1145 (2018).
Zhang, Y. et al. IsomiR Bank: a research resource for tracking IsomiRs. Bioinformatics 32, 2069–2071 (2016).
Dong, H., Nilsson, L. & Kurland, C. G. Co-variation of tRNA abundance and codon usage in Escherichia coli at differing growth rates. J. Mol. Biol. 260, 649–663 (1996).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
Grant, S. S. & Hung, D. T. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 4, 273–283 (2013).
Zhang, Y. Persisters, persistent infections and the Yin–Yang model. Emerg. Microbes Infect. 3, e3 (2014).
Steinberg, S., Misch, A. & Sprinzl, M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 21, 3011–3015 (1993).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016).
Chionh, Y. H. et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7, 13302 (2016).
Doyle, F. et al. Gene- and genome-based analysis of significant codon patterns in yeast, rat and mice genomes with the CUT Codon UTilization tool. Methods 107, 98–109 (2016).
Phizicky, E. M. & Hopper, A. K. tRNA biology charges to the front. Genes Dev. 24, 1832–1860 (2010).
Chernyakov, I., Whipple, J. M., Kotelawala, L., Grayhack, E. J. & Phizicky, E. M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5’-3’ exonucleases Rat1 and Xrn1. Genes Dev. 22, 1369–1380 (2008).
Hopper, A. K., Pai, D. A. & Engelke, D. R. Cellular dynamics of tRNAs and their genes. FEBS Lett. 584, 310–317 (2010).
Anderson, P. & Ivanov, P. tRNA fragments in human health and disease. FEBS Lett. 588, 4297–4304 (2014).
Cruz, J. W. et al. Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat. Commun. 6, 7480 (2015).
Schifano, J. M. et al. tRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 44, 1256–1270 (2016).
Lyons, S. M., Fay, M. M. & Ivanov, P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett. 592, 2828–2844 (2018).
Fu, H. et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583, 437–442 (2009).
Haiser, H. J., Karginov, F. V., Hannon, G. J. & Elliot, M. A. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 36, 732–741 (2008).
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018).
Kietrys, A., Velema, W. & Kool, E. Fingerprints of modified RNA bases from deep sequencing profiles. J. Am. Chem. Soc. 139, 17074–17081 (2017).
Hauenschild, R. et al. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 43, 9950–9964 (2015).
Levanon, E. Y. et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22, 1001–1005 (2004).
Elenbaas, B. et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 15, 50–65 (2001).
Kendall, S. D., Adam, S. J. & Counter, C. M. Genetically engineered human cancer models utilizing mammalian transgene expression. Cell Cycle 5, 1074–1079 (2006).
Qattan, A. et al. Robust expression of tumor suppressor miRNA’s let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer 17, 799 (2017).
Xuan, P., Li, L., Zhang, T., Zhang, Y. & Song, Y. Prediction of disease-related microRNAs through integrating attributes of microRNA nodes and multiple kinds of connecting edges. Molecules 24, 3099 (2019).
Wang, X. et al. Differential expression profile analysis of miRNAs with HER-2 overexpression and intervention in breast cancer cells. Int. J. Clin. Exp. Pathol. 10, 5039–5062 (2017).
Maltseva, D. V. et al. miRNome of inflammatory breast cancer. BMC Res. Notes 7, 871 (2014).
Ueda, S., Takanashi, M., Sudo, K., Kanekura, K. & Kuroda, M. miR-27a ameliorates chemoresistance of breast cancer cells by disruption of reactive oxygen species homeostasis and impairment of autophagy. Lab. Invest. 100, 863–873 (2020).
Pirouz, M., Ebrahimi, A. G. & Gregory, R. I. Unraveling 3’-end RNA uridylation at nucleotide resolution. Methods 155, 10–19 (2019).
Sorefan, K. et al. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence 3, 4 (2012).
Dard-Dascot, C. et al. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics 19, 118 (2018).
Wong, R. K. Y., MacMahon, M., Woodside, J. V. & Simpson, D. A. A comparison of RNA extraction and sequencing protocols for detection of small RNAs in plasma. BMC Genomics 20, 446 (2019).
Heinicke, F. et al. Systematic assessment of commercially available low-input miRNA library preparation kits. RNA Biol. 17, 75–86 (2020).
Chu, Y. et al. Intramolecular circularization increases efficiency of RNA sequencing and enables CLIP-Seq of nuclear RNA from human cells. Nucleic Acids Res. 43, e75 (2015).
Lucigen CircLigase II Product Manual, p. 3. https://www.lucigen.com/docs/manuals/MA298E-CircLigase-II-ssDNA-Ligase.pdf (July 2019).
Xiong, Y. et al. A comparison of mRNA sequencing with random primed and 3’-directed libraries. Sci. Rep. 7, 14626 (2017).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Hia, F. et al. Mycobacterial RNA isolation optimized for non-coding RNA: high fidelity isolation of 5S rRNA from Mycobacterium bovis BCG reveals novel post-transcriptional processing and a complete spectrum of modified ribonucleosides. Nucleic Acids Res. 43, e32 (2015).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank members of D. Bartel’s laboratory (Whitehead Institute and MIT Department of Biology), especially A. Stefano and D. Bartel, for assistance with RNA blots. We thank P. Ivanov (Harvard Medical School) for sharing synthetic RNA standards. This work was supported by grants from the National Natural Science Foundation of China (no. 32070629 to B.C.), the US National Science Foundation (no. MCB-1412379 to V.C.-L.), the National Institute of Environmental Health Sciences (no. ES002109) and the National Research Foundation of Singapore through the Singapore-MIT Alliance for Research and Technology Antimicrobial Resistance IRG (P.C.D.). J.F.H. was supported by MIT Toxicology Training grant no. T32-ES007020, D.Y. by a postdoctoral fellowship from A*STAR, Singapore and S.M.H. by a postdoctoral fellowship from the Swiss National Science Foundation.
Author information
Authors and Affiliations
Contributions
P.C.D., B.C. and J.F.H. conceived of AQRNA-seq, designed the experiments and wrote the first draft of the manuscript. P.C.D., J.F.H., B.C. and D.Y. developed the method and performed the sequencing experiments. J.F.H., B.C., D.M. and S.S.L. developed, implemented and interpreted the data-processing workflows and computational analyses. D.Y., S.V. and J.F.H. performed mycobacterial culturing and RNA isolation. T.J.B. analyzed proteomics data for codon usage patterns. J.M.B. performed E. coli culturing and RNA isolation. N.D. performed proteomics analyses. S.M.H. optimized experimental conditions and characterized demethylation efficiency by LC–MS. S.M.H. and J.F.H. performed RNA blot analyses. M.S.D. contributed reagents and analyzed miRNA data. J.Z. analyzed miRNA data. V.C.-L. supervised E. coli experiments and contributed insights and analysis. All authors participated in the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
B.C., J.F.H., D.Y., S.M.H., M.S.D. and P.C.D. are co-inventors on two patents (PCT/US2019/013714, US 2019/0284624 A1) relating to the published work.
Additional information
Peer review information Nature Biotechnology thanks James Hadfield and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, Tables 1–3, Discussion and References.
Rights and permissions
About this article
Cite this article
Hu, J.F., Yim, D., Ma, D. et al. Quantitative mapping of the cellular small RNA landscape with AQRNA-seq. Nat Biotechnol 39, 978–988 (2021). https://doi.org/10.1038/s41587-021-00874-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-00874-y
This article is cited by
-
Small RNA structural biochemistry in a post-sequencing era
Nature Protocols (2024)
-
Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing
Nature Biotechnology (2024)
-
Roles and regulation of tRNA-derived small RNAs in animals
Nature Reviews Molecular Cell Biology (2024)
-
The role and mechanism of action of tRNA-derived fragments in the diagnosis and treatment of malignant tumors
Cell Communication and Signaling (2023)
-
Small RNA modifications: regulatory molecules and potential applications
Journal of Hematology & Oncology (2023)