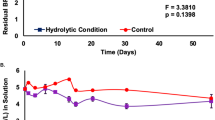
Abstract
The explosive hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), a major component of munitions, is used extensively on military training ranges. As a result, widespread RDX pollution in groundwater and aquifers in the United States is now well documented. RDX is toxic, but its removal from training ranges is logistically challenging, lacking cost-effective and sustainable solutions. Previously, we have shown that thale cress (Arabidopsis thaliana) engineered to express two genes, xplA and xplB, encoding RDX-degrading enzymes from the soil bacterium Rhodococcus rhodochrous 11Y can break down this xenobiotic in laboratory studies. Here, we report the results of a 3-year field trial of XplA/XplB-expressing switchgrass (Panicum virgatum) conducted on three locations in a military site. Our data suggest that XplA/XplB switchgrass has in situ efficacy, with potential utility for detoxifying RDX on live-fire training ranges, munitions dumps and minefields.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors on request.
References
Field, S. Q. Boom!: The Chemistry and History of Explosives (Chicago Review Press, 2017).
BCC Research LLC. Global market for explosives to gain $7.4 billion from 2017–2022. https://www.bccresearch.com/pressroom/chm/global-market-for-explosives-to-gain-$74-billion-from-2017-2022 (2018).
United States Environmental Protection Agency. Technical fact sheet – hexahydro-1,3,5-trinitro1,3,5-triazine (RDX). http://www.epa.gov/sites/production/files/2017-10/documents/ffrro_ecfactsheet_rdx_9-15-17_508.pdf (2017).
Rylott, E. L., Lorenz, A. & Bruce, N. C. Biodegradation and biotransformation of explosives. Curr. Opin. Biotechnol. 22, 434–440 (2011).
Rylott, E. L. & Bruce, N. C. Right on target: using plants and microbes to remediate explosives. Int. J. Phytoremediation 21, 1051–1064 (2019).
Toghiani, R. K., Toghiani, H., Maloney, S. W. & Boddu, V. M. Prediction of physicochemical properties of energetic materials. Fluid Phase Equilib. 264, 86–92 (2008).
Yamamoto, H., Morley, M. C., Speitel, G. E. & Clausen, J. Fate and transport of high explosives in a sandy soil: adsorption and desorption. Soil Sediment Contam. 13, 361–379 (2004).
Clausen, J., Robb, J., Curry, D. & Korte, N. A case study of contaminants on military ranges: Camp Edwards, Massachusetts, USA. Environ. Pollut. 129, 13–21 (2004).
Albright, R. Cleanup of Chemical and Explosive Munitions. (Elsevier, 2012).
Pichtel, J. Distribution and fate of military explosives and propellants in soil: a review. Appl. Environ. Soil Sci. 2012, 617236 (2012).
United States General Accounting Office. Department of Defense Operational Ranges: More Reliable Cleanup Cost Estimates and a Proactive Approach to Identifying Contamination Are Needed (GAO, 2004).
Kennen K. & Kirkwood, N. Phyto: Principles and Resources for Site Remediation and Landscape Design 1st edn (Routledge, 2015).
Jackson, R., Rylott, E., Fournier, D., Hawari, J. & Bruce, N. Exploring the biochemical properties and remediation applications of the unusual explosive-degrading P450 system XpIA/B. Proc. Natl Acad. Sci. USA 104, 16822–16827 (2007).
Rylott, E. L. et al. An explosive-degrading cytochrome P450 activity and its targeted application for the phytoremediation of RDX. Nat. Biotechnol. 24, 216–219 (2006).
Seth-Smith, H. M. et al. Cloning, sequencing, and characterization of the hexahydro-1,3,5-trinitro-1,3,5-triazine degradation gene cluster from Rhodococcus rhodochrous. Appl. Environ. Microbiol. 68, 4764–4771 (2002).
Paquet, L., Monteil-Rivera, F., Hatzinger, P. B., Fuller, M. E. & Hawari, J. Analysis of the key intermediates of RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine) in groundwater: occurrence, stability and preservation. J. Environ. Monit. 13, 2304–2311 (2011).
Bhadra, R. et al. Studies on plant-mediated fate of the explosives RDX and HMX. Chemosphere 44, 1259–1264 (2001).
Van Aken, B., Yoon, J. M., Just, C. L. & Schnoor, J. L. Metabolism and mineralization of hexahydro-1,3,5-trinitro-1,3,5-triazine inside poplar tissues (Populus deltoides x nigra DN-34). Environ. Sci. Technol. 38, 4572–4579 (2004).
Just, C. L. & Schnoor, J. L. Phytophotolysis of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) in leaves of reed canary grass. Environ. Sci. Technol. 38, 290–295 (2004).
Rylott, E. L., Johnston, E. J. & Bruce, N. C. Harnessing microbial gene pools to remediate persistent organic pollutants using genetically modified plants—a viable technology? J. Exp. Bot. 66, 6519–6533 (2015).
Zhang, L. et al. Expression in grasses of multiple transgenes for degradation of munitions compounds on live-fire training ranges. Plant Biotechnol. J. 15, 624–633 (2017).
Hewitt, A. D., Jenkins, T. F., Walsh, M. E., Walsh, M. R. & Taylor, S. RDX and TNT residues from live-fire and blow-in-place detonations. Chemosphere 61, 888–894 (2005).
Schneider, U. A. & McCarl, B. A. Economic potential of biomass based fuels for greenhouse gas emission mitigation. Environ. Resour. Econ. 24, 291–312 (2003).
Fournier, D., Trott, S., Hawari, J. & Spain, J. Metabolism of the aliphatic nitramine 4-nitro-2,4-diazabutanal by Methylobacterium sp. strain JS178. Appl. Environ. Microbiol. 71, 4199–4202 (2005).
Zhang, L., Rylott, E. L., Bruce, N. C. & Strand, S. E. Genetic modification of western wheatgrass (Pascopyrum smithii) for the phytoremediation of RDX and TNT. Planta 249, 1007–1015 (2018).
Rylott, E. L. et al. The explosive-degrading cytochrome P450 XplA: biochemistry, structural features and prospects for bioremediation. Biochim. Biophys. Acta 1814, 230–236 (2011).
Alexandrova, K. S., Denchev, P. D. & Conger, B. V. Micropropagation of switchgrass by node culture. Crop Sci. 36, 1709–1711 (1996).
EPA. Method 8330A: nitroaromatics and nitramines by high performance liquid chromatography (HPLC). https://www.epa.gov/sites/production/files/2015-12/documents/8330a.pdf (2007).
Larson, S. L., Strong, A. B., Yost, S. L., Escalon, B. L. & Parker D. Analysis of Explosives in Plant Tissues: Modifications to Method 8330 for Soil. Technical report IRRP-98-5 (US Army Corps of Engineers, 1998).
Dunn, O. J. Multiple comparisons using rank sums. Technometrics 6, 241–252 (1964).
Acknowledgements
We thank N. Podpora, H. Kase and Z. Pick for maintenance and monitoring of the field plots and T. Lent and I. Warden from Fort Drum Directorate of Plans, Training, Mobilization and Security, Range Branch, Fort Drum, New York. This work was funded by the Strategic Environmental Research and Development Program (ER-1498) and Environmental Security Technology Certification Program (ER-201436) of the US Department of Defense. The trial was conducted under APHIS permit number 16–025–103rm.
Author information
Authors and Affiliations
Contributions
T.J.C., E.L.R., A.J.P., S.E.S. and N.C.B. conceived the study and designed the experiments. T.J.C. was responsible for plot design, construction and sample collection. T.J.C., L.Z. and R.M.R. performed biomass and RDX quantification. E.L.R., T.J.C., S.E.S., L.Z. and N.C.B. analyzed the data. E.L.R. performed statistical analyses, and L.Z. performed all other experiments. E.L.R. and N.C.B. wrote the manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biotechnology thanks Neal Stewart and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Map of explosives contamination across the globe.
Country names link to source information. Stars indicate documented RDX contamination, with details listed in Supplementary Table 1.
Extended Data Fig. 2 Design of the field plots and layout.
(a) Randomized block lay-out of the plots. (b) Location of the plant lines on each plot. (c) Location of each of the three, independently-transformed XplAB-expressing lines N1, N2 and N5. (d) Aerial photograph of the plot taken September 2015 before planting.
Extended Data Fig. 3
Appearance of the plants at the end of each growing season over the three-year trial.
Extended Data Fig. 4 Aerial tissue weights following harvest at the end of the field trial.
(a) Biomass of the WT (wild type) and individual (TG) transgenic N lines on uncontaminated and RDX-treated plots (n = 45(WT), n = 15(TG) plants). Statistical significance was performed using one-way analysis of variance followed by Tukey’s post hoc test. On No RDX plots, line N5 mean biomass was significantly (P < 0.004) lower than N2. (b) Comparison of biomass from individual plots (n = 15 plants). A Kruskal-Wallis H test showed the distributions of biomass values for the No RDX plots were statistically significantly different (P = 0.002). Pairwise comparisons were performed using Dunn’s (1964) procedure with a Bonferroni correction for multiple comparisons, and presenting adjusted, two-sided P values. This post hoc analysis revealed that the plant biomass from TG plot 12 was statistically significantly different from TG plots 21 and 26 (P = 0.013). (c) Total biomass for each of the treatment types WT and transgenic (TG) plants (n = 3 plots). For figures a, b and c, the top and bottom box lines show the first and third quartiles, the line in the middle of each box is the median. The whiskers show the maximum and minimum values, with the exceptions of outliers (circles) and extremes (asterisks). Outliers are at least 1.5 box lengths from the median, and extremes are at least 3 box lengths from the median.
Extended Data Fig. 5 Gene expression analysis on leaf tissue harvested from the plots in September 2018.
(a) Transcript abundance measured using quantitative RT- PCR for xplA and (b) xplB expression. Conditions were as described earlier55, with transcripts normalized to the switchgrass reference gene eIF-4a. Error bars represent mean ± SE, n = 12 of triplicate measurements on single wild type (WT) plants from each of plots 4, 7 and13; and triplicate measurements on single plants from plots 2, 23 and 26 for each transgenic line. A two-tailed t-test showed the transgenic lines N1, N2 and N5 were significantly different (*) from WT (for xplA, P = < 0.00000 for N1, 0.00029 for N2, and 0.02198 for N5; for xplB, P = < 0.00000 for N1 and N2, P = 0.00018 for N5). (c) Western blot analysis conducted as described earlier55 using XplA antibody56 and RuBisCO large subunit antibody (AS03 037, Agrisera, Sweden). Band intensities for each line, and antibody, were quantified using ImageJ software57. Error bars represent ± SE, n = 8 replicate blots. A two-tailed t-test showed the transgenic lines N1, N2 and N5 were significantly different (*) from WT (P = 0.00018, 0.00008 and 0.00002 respectively).
Extended Data Fig. 6 Total tissue RDX from plots over the 2018 growing season.
Mean aerial tissue RDX concentration in plants from the 100 ppm RDX plots (ND, Not Detected, n = 45). The top and bottom box lines show the first and third quartiles, the line in the middle of each box is the median. The whiskers show the maximum and minimum values, with the exceptions of outliers (circles) and extremes (asterisks). Outliers are at least 1.5 box lengths from the median, and extremes are at least 3 box lengths from the median.
Extended Data Fig. 7 RDX soil, soil water and tank concentrations in 2018 growing season.
(a) Soil RDX concentration in the 100 ppm plots measured at three depths at the start (April n= 9 soil samples) and end (Oct n= 27 soil samples) of the growing season (NPC; No plant control, WT; wild type, TG, transgenic. (b) Heat plot showing mean distribution of RDX across the 100 ppm plots at the three depths, c = position of culvert, tank = position of water tank. (c) Concentration of RDX in lysimeters and tanks across the 2018 growing season. Lysimeter data: n = 12(NPC), 15(WT), 15(TG). Tank data: n = 11(NPC), 12(WT), 12(TG). For the tank data, a Kruskal-Wallis H test showed the distributions of NPC, WT and TG values were statistically significantly different (P = 0.026) from each other. Pairwise comparisons were performed using Dunn’s (1964) procedure with a Bonferroni correction for multiple comparisons, and presenting adjusted, two-sided P values. This post hoc analysis revealed that the RDX concentrations in the TG tanks were statistically significantly different from those in the NPC tanks (P = 0.021). For figures a and c, The top and bottom box lines show the first and third quartiles, the line in the middle of each box is the median. The whiskers show the maximum and minimum values, with the exceptions of outliers (circles) and extremes (asterisks). Outliers are at least 1.5 box lengths from the median, and extremes are at least 3 box lengths from the median.
Extended Data Fig. 8 Excess water pumped from plots over each growing season.
Volume of excess water pumped from the plots over each growing season of the field trial. (a) No RDX plots. (b) 100ppm RDX plots. (NPC; No plant control, WT; wild type, TG, transgenic, data shown for each graphs are mean ± SD, n = 3 plots). Letters denote values that were significantly between the three treatment types for that growing season. For the 2017 No RDX data, a Kruskal-Wallis H test showed the distributions of NPC, WT and TG values were statistically significantly different (P = 0.014) from each other. Pairwise comparisons were performed using Dunn’s (1964) procedure with a Bonferroni correction for multiple comparisons. For all over data, one-way analysis of variance followed by Tukey’s post hoc test was used, with P<0.05 considered as significant.
Extended Data Fig. 9 Leaf surface area of the plants at the end of the 2018 growing season.
(a) Mean leaf surface areas of the wild type (WT) and individual transgenic (TG) lines grown in the laboratory (n = 3 plants). (b) Comparison of mean leaf surface area of all WT and TG plants grown on the uncontaminated (No RDX) verses RDX-treated (+ RDX) plots (n = 6 plots) (c) Mean leaf surface areas in the WT and individual TG, N lines grown in the field, independent of the presence of RDX (n = 3 plots). (d) Mean leaf surface areas in the WT and TG lines grown in uncontaminated verses RDX-treated (n = 3 for WT, n= 9 for TG). For all figures, the top and bottom box lines show the first and third quartiles, the line in the middle of each box is the median. The whiskers show the maximum and minimum values.
Extended Data Fig. 10 Leaf transpiration rates.
Leaf surface transpiration rates for wild type and transgenic line N1 from adaxial and abaxial surfaces (Error bars represent mean ± SD, n = 20 leaves).
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Cary, T.J., Rylott, E.L., Zhang, L. et al. Field trial demonstrating phytoremediation of the military explosive RDX by XplA/XplB-expressing switchgrass. Nat Biotechnol 39, 1216–1219 (2021). https://doi.org/10.1038/s41587-021-00909-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-00909-4
This article is cited by
-
The efficient applications of native flora for phytorestoration of mine tailings: a pan-global survey
Environmental Science and Pollution Research (2024)
-
Sustainable remediation and redevelopment of brownfield sites
Nature Reviews Earth & Environment (2023)
-
Safety management regulation and practice standards on living modified organism (LMO) facilities under the Ministry of Environment
Plant Biotechnology Reports (2023)
-
Characterization of diazotrophic root endophytes in Chinese silvergrass (Miscanthus sinensis)
Microbiome (2022)