Abstract
Chromatin is organized into topologically associating domains (TADs) enriched in distinct histone marks. In cancer, gain-of-function mutations in the gene encoding the enhancer of zeste homolog 2 protein (EZH2) lead to a genome-wide increase in histone-3 Lys27 trimethylation (H3K27me3) associated with transcriptional repression. However, the effects of these epigenetic changes on the structure and function of chromatin domains have not been explored. Here, we found a functional interplay between TADs and epigenetic and transcriptional changes mediated by mutated EZH2. Altered EZH2 (p.Tyr646* (EZH2Y646X)) led to silencing of entire domains, synergistically inactivating multiple tumor suppressors. Intra-TAD gene silencing was coupled with changes of interactions between gene promoter regions. Notably, gene expression and chromatin interactions were restored by pharmacological inhibition of EZH2Y646X. Our results indicate that EZH2Y646X alters the topology and function of chromatin domains to promote synergistic oncogenic programs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
In this study, we used the following mRNA expression datasets: GSE23501 for wild-type and EZH2-mutated GCB-DLBCL primary human samples, PRJNA278311 (NCBI-BioProject) for wild-type and EZH2-mutated FL primary human samples, GSE40792 for wild-type and EZH2-mutated cell lines before and after treatment with GSK126, GSE49284 for EZH2-mutated cell lines before and after treatment with EPZ6438, and GSE12195 for centrocytes, centroblasts, and memory B cells. ChIP-seq data for H3K27me3 in OCI-Ly7, DOHH-2, and Karpas-422 were downloaded from ENCODE; H3K27me3 in WSU-DLCL2 was downloaded from GSE40970; H3K4me3 in OCI-Ly7 and Karpas-422 were downloaded from ENCODE. ChIP-seq data for H3K27me3 and RNA-seq data for OCI-Ly19 and OCI-Ly19-EZH2Y646F were generated as described in the manuscript and have been deposited at GSE114270. HiC matrices and UMI-4C data have been deposited at Zenodo: https://doi.org/10.5281/zenodo.1244182. Custom scripts are available through a public GitHub repository at: https://github.com/CSOgroup/Donaldson-et-al-scripts/.
References
Bonev, B. & Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016).
Dixon, J. R., Gorkin, D. U. & Ren, B. Chromatin domains: the unit of chromosome organization. Mol. Cell 62, 668–680 (2016).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Le Dily, F. et al. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 28, 2151–2162 (2014).
de Laat, W. & Duboule, D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502, 499–506 (2013).
Fraser, J. et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 11, 852 (2015).
Hnisz, D. et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods.Science 351, 1454–1458 (2016).
Weischenfeldt, J. et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74 (2017).
Flavahan, W. A. et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016).
Taberlay, P. C. et al. Three-dimensional disorganisation of the cancer genome occurs coincident with long range genetic and epigenetic alterations. Genome Res. 26, 719–731 (2016).
Plass, C. et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 14, 765–780 (2013).
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 42, 181–185 (2010).
Souroullas, G. P. et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat. Med. 22, 632–640 (2016).
Tirode, F. et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 4, 1342–1353 (2014).
Comet, I., Riising, E. M., Leblanc, B. & Helin, K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 16, 803–810 (2016).
Sneeringer, C. J. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA 107, 20980–20985 (2010).
Yap, D. B. et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459 (2011).
Béguelin, W. et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013).
Oricchio, E. et al. Genetic and epigenetic inactivation of SESTRIN1 controls mTORC1 and response toEZH2 inhibition in follicular lymphoma. Sci. Transl. Med. 9, eaak9969 (2017).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Yang, T. et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 27, 1939–1949 (2017).
Pfitzner, D., Leibbrandt, R. & Powers, D. Characterization and evaluation of similarity measures for pairs of clusterings. Knowl. Inf. Syst. 19, 361 (2009).
Shin, H. et al. TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res. 44, e70 (2016).
Zufferey, M., Tavernari, D., Oricchio, E. & Ciriello, G. Comparison of computational methods for the identification of topologically associating domains. Genome. Biol. 19, 217 (2018).
Carty, M. et al. An integrated model for detecting significant chromatin interactions from high-resolution Hi-C data. Nat. Commun. 8, 15454 (2017).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Ortega-Molina, A. et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat. Med. 21, 1199–1208 (2015).
Nuytten, M. et al. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene 27, 1449–1460 (2008).
Klein, U. et al. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl Acad. Sci. 100, 2639–2644 (2003).
Yang, C.-S. et al. Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol. Cell 66, 503–516.e5 (2017).
Matloubian, M. et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360 (2004).
Dansen, T. B. & Burgering, B. M. T. Unravelling the tumor-suppressive functions of FOXO proteins. Trends. Cell Biol. 18, 421–429 (2008).
Okosun, J. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46, 176–181 (2014).
Oricchio, E. et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell 147, 554–564 (2011).
Chen, B. B., Glasser, J. R., Coon, T. A. & Mallampalli, R. K. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene 31, 2566–2579 (2012).
Li, L., Pan, D., Chen, H., Zhang, L. & Xie, W. F-box protein FBXL2 inhibits gastric cancer proliferation by ubiquitin-mediated degradation of forkhead box M1. FEBS Lett. 590, 445–452 (2016).
Hatzimichael, E. et al. The collagen prolyl hydroxylases are novel transcriptionally silenced genes in lymphoma. Br. J. Cancer 107, 1423–1432 (2012).
Chambwe, N. et al. Variability in DNA methylation defines novel epigenetic subgroups of DLBCL associated with different clinical outcomes. Blood 123, 1699–1708 (2014).
Oricchio, E. et al. Frequent disruption of the RB pathway in indolent follicular lymphoma suggests a new combination therapy. J. Exp. Med. 211, 1379–1391 (2014).
Mavrakis, K. J. et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 12, 372–379 (2010).
Scuoppo, C. et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature 487, 244–248 (2012).
Schatz, J. H. et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J. Exp. Med. 208, 1799–1807 (2011).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015).
Dao, L. T. M. et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 49, 1073–1081 (2017).
Boettiger, A. N. et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Budanov, A. V. & Karin, M. The p53-regulated Sestrin gene products inhibit mTOR signaling. Cell 134, 451–460 (2008).
Renault, V. M. et al. The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor. Oncogene 30, 3207–3221 (2011).
Schwartzman, O. et al. UMI-4C for quantitative and targeted chromosomal contact profiling. Nat. Methods 13, 685–691 (2016).
Kleinberg, J. & Tardos, É. Algorithm Design (Pearson, Boston, 2005).
Acknowledgements
We thank B. Ren and A. D. Schmitt for help with Hi-C library preparation; C. Bolt for help with the UMI-4C protocol; J. Lingner, D. Trono, and F. Radtke for critical reading of the manuscript; and D. Duboule and J. Huelsken for useful discussions. We thank the EPFL research animal, flow cytometry, histology, and sequencing facilities. This work is supported by the ISREC Foundation (E.O.), the Swiss National Science Foundation (E.O. and M.C.D.-C. SNF-31003A_159637) and Swiss Cancer League (E.O. KFS-3982-08-2016). G.C. is supported by the Giorgi-Cavaglieri Foundation. D.T. is supported by the Swiss National Science Foundation (SNSF, SNF-310030_169519), M.Z. is supported by the Swiss Cancer League (KFS-3983-08-2016), and S.S. is supported by European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 665667.
Author information
Authors and Affiliations
Contributions
M.C.D-C. prepared Hi-C, UMI-4C, ChIP-seq and RNA-seq libraries and performed in vitro validation experiments and DNA FISH experiments. S.S. analyzed Hi-C, UMI-4C, and ChIP-seq data. M.Z. performed the comparison of Hi-C contact maps, TAD calling, and all analyses based on mRNA expression data. D.T. performed interactome analyses and all analyses based on ChIP-seq data. K.M.D. and T.R. acquired and analyzed STORM images. N.K. and E.B. performed in vitro and in vivo experiments. M.M. assisted in the analysis of Hi-C data. F.R. assisted in the analysis of STORM data. S.M. supervised in STORM acquisition and image analyses. G.C and E.O. designed the study and wrote the manuscript with comments from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
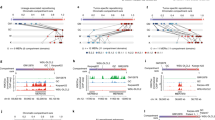
Supplementary Figure 1 Detailed comparison of the 3D genome organization in EZH2WT and EZH2Y646X cells.
a, Immunoblot of H3K27me3, H3K27me2, and Histone-3 in EZH2 wild-type (WT) and EZH2 mutated (Y646X) cells (n = 2 experiments). b-e, Comparisons of intra-chromosomal maps (n = 22 maps in each comparison) of EZH2Y646X cell lines (Karpas422 and WSU-DLCL2) and of the indicated cell lines based on (b) stratum-adjusted correlation coefficient (SCC), (c) fraction of 1Mb bins assigned to different compartments, (d) TAD similarity by Measure of Concordance, and (e) fraction of shared significant interactions. The thick central line of each box plot represents the median, the bounding box corresponds to the 25th–75th percentiles, and the whiskers extend up to 1.5 times the interquartile range, f, Fraction of RNA-seq reads mapping to EZH2WT and EZH2Y646F in OCI-Ly19 cells in three independent replicates (Rep1, Rep2, Rep3). g, Immunoblot of H3K27me3, Histone-3, EZH2, and tubulin in OCI-Ly19 EZH2 wild-type (WT), OCI-Ly19 EZH2 wild-type transduced with the vector, and OCI-Ly19 EZH2Y646F cells. h, Comparisons of intra-chromosomal Hi-C matrices (n = 22 maps per comparison) between OCI-Ly19-EZH2Y646F cells and the indicated cell lines by: SCC, fraction of 1Mb bins assigned to different compartments, TAD similarity, and fraction of shared significant interactions. The thick central line of each box plot represents the median, the bounding box corresponds to the 25th–75th percentiles, and the whiskers extend up to 1.5 times the interquartile range.
Supplementary Figure 2 H3K27me3 across subcompartments and TADs in EZH2WT and EZH2Y646F cells.
a-b, Distribution of (a) mean H3K27me3 and (b) mean H3K27me3 fold-changes between OCI-Ly19-EZH2Y646F and OCI-Ly19-EZH2WT in regions within sub-compartments A1 (n = 490), A2 (n = 1249), B1 (n = 896), B2 (n = 504), B3 (n = 685), and B4 (n = 25) in OCI-Ly19-EZH2WT and OCI-Ly19-EZH2Y646F. The thick central line of each box plot represents the median value, the bounding box corresponds to the 25th–75th percentiles, and the whiskers extend up to 1.5 times the interquartile range. Dots are values extending beyond such range. c, Pearson’s correlation between TAD mean H3K27me3 in OCI-Ly19-EZH2Y646F and the mean of the corresponding values in Karpas-422 and WSU-DLCL2 (n = 2,038 TADs). d-e, Correlation across n = 2,038 TADs of H3K27me3 in (d) OCI-Ly19-EZH2WT and (e) OCI-Ly19-EZH2Y646F in loci within the same TAD and loci separated by one TAD boundary. f, Distribution of TAD mean H3K27me3 fold-changes (log2FC) within = 2,038 TADs in OCI-Ly19 EZH2Y646F versus OCI-Ly19 EZH2WT compared to fold-change distributions obtained after permuting H3K27me3 bins (50kb or 100kb) preserving the compartment composition. g, Ratio between the expected number of TADs with >2-fold H3K27me3 increase in OCI-Ly19-EZH2Y646F compared to OCI-Ly19-EZH2WT (log2(FC) > 1, Y-axis) based on permutation of H3K27me3 bins of sizes ranging from 50 kb to 2 Mb (interval size is on the X-axis) and the observed number of TADs with log2(FC) > 1.
Supplementary Figure 3 Transcriptionally inactive TADs in EZH2WT and EZH2Y646X cells.
a, Formula of mRNA Fold-Change Concordance (FCC) score and graphical representation of the cumulative sum curves used to quantify the concordance of gene expression fold-changes within TADs. b, Cumulative sum curve of mRNA FCC scores derived from the comparison of OCI-Ly19-EZH2Y646F versus OCI-Ly19-EZH2WT cells in n = 900 TADs. c-e, Pearson’s correlation of mRNA expression profiles (Y-axis) between gene-pairs located in the same TAD and in different TADs as a function of their genomic distance (X-axis) in (c) DLBCL cell lines (n = 16), (d) GC-DLBCL patient samples (n = 37), and (e) FL patient samples (n = 23). Curves were fitted with the loess R function. f, Comparison of TAD mean H3K27me3 fold-changes (X-axis) versus the TAD mean mRNA expression fold-changes (Y-axis) in OCI-Ly19-EZH2Y646F versus OCI-Ly19-EZH2WT cells (n = 900 TADs). Points are colored to reflect point density: cold colors indicate low density; warm colors indicate high density. Highlighted areas correspond to regions with a >2-fold difference in both H3K27me3 and mRNA expression. g-h, Comparison of TAD mean H3K27me3 fold-changes (X-axis) versus the TAD mean mRNA expression fold-changes (Y-axis) in OCI-Ly19-EZH2Y646F versus OCI-Ly19-EZH2WT cells considering only TADs including genes exhibiting a mRNA expression fold change |log2 FC| >1 (n = 187 TADs), or an adjusted p-value < 0.1 (n = 166 TADs). Only TADs with at least 3 genes are considered. i, (Left) Overlap between 37 out of 72 inactive TADs containing at least 3 genes with detectable expression in OCI-Ly19-EZHY646F and OCI-Ly19-EZHWT and 89 inactive TADs defined based on H3K27me3 and mRNA fold-changes in OCI-Ly19. (Right): Overlap of 72 inactive TADs with 57 inactive TADs defined based on H3K27me3 fold-changes obtained by comparing EZH2 mutated cell lines (Karpas-422 and WSU-DLCL2) and OCI-Ly19 EZH2WT. P-value determined by two-tailed Fisher’s exact test. OR: odd ratios. j, Mean mRNA expression difference in inactive TADs in normal centroblast and centrocytes (n = 10 samples of germinal center, GC, cells) versus memory B-cells (n = 5 samples, X-axis) and corresponding p-value determined by two-tailed T-test. k, mRNA expression heatmap of genes in the top 3 differentially expressed inactive TADs in centroblast/centrocytes versus memory B-cells.
Supplementary Figure 4 Inactive TADs downregulate tumor suppressors.
a, Quantification of H3K27me3 by ChIP-qPCR for genes included in TAD6.139 (FOXO3, n = 3 and SESN1, n = 2) in Toledo (EZH2WT) and WSU-DLCL2 (EZH2Y646F) cells. Data were normalized based on the input, and chromatin immune precipitation of Histone-3 (H3-ChIP) was used as control. Bars indicate mean values and error bars correspond to 1 standard deviation. b, Quantitative mRNA expression analysis of genes included in TAD6.139 (FOXO3, ARMC2, and SESN1) in OCI-Ly19 EZH2WT (n = 3 independent experiments) compared to OCI-Ly19 expressing empty vector (pLVX) (n = 1) or OCI-Ly19 EZH2Y646F (n = 3). Data are normalized to GAPDH. Error bars correspond to 1 standard deviation. P-values were calculated using a two-tailed t-test. c, Hi-C contact map at 20kb resolution of the genomic region Chr.3 32.7-33.8Mb in OCI-Ly19 EZH2WT. TAD boundaries are contoured in black and representative genes within TADs are shown. d, H3K27me3 ChIP-seq tracks (n = 3 experiments) on Chr.3 32.7-33.8Mb in OCI-Ly19 EZH2WT and OCI-Ly19 EZH2Y646. e, Barplot showing mRNA expression fold-changes of individual genes in Chr.3 32.7-33.8Mb between OCI-Ly19-EZH2Y646F and OCI-Ly19-EZH2WT and between n = 12 EZH2Y646X cell lines and n = 4 EZH2WT cell lines.
Supplementary Figure 5 Synergistic downregulation of genes in TAD6.139 promotes lymphomagenesis.
a-c, Differential expression analyses in FL5-12 pro-B cells transduced with vector control or shRNAs targeting (a) Sesn1, (b) Foxo3, or (c) Armc2 in three independent experiments. Bars indicate mean values; error bars correspond to 1 standard deviation. P-values were calculated using a two-tailed t-test. d, Percentage of cells in each sub-population transduced with control vectors at day 4, 8, and 12 relatives to day 0. Mean values (bars) and standard deviations (error bars) were calculated based on 2 independent experiments. n.i.: no infected cells. e-f, Percentage of cells in each sub-population transduced with the indicated shRNAs at day 4, 8, and 12 relatives to day 0. Mean values (bars) and standard deviations (error bars) were calculated based on 2 independent experiments. n.i.: no infected cells. g, Overall survival of lethally irradiated animals transplanted with Eμ-myc HPC expressing vector (n = 14 animals), shFoxo3 (n = 16), shSesn1 (n = 17), or mix population of shSesn1 and shFoxo3 (n = 21). P-values were calculated by Log-rank (Mantel-Cox) test for each population versus the vector. h, Analysis by flow cytometry of CD45-R and CD3 expression in the indicated tumours (n = 2 experiments). i, Quantification of the CD45-R and CD3 positive cells in the indicated tumours.
Supplementary Figure 6 Differential expression analysis after treatment with EZH2 inhibitors leads to TAD reactivation.
a-b, Distributions of gene expression fold-changes between EZH2Y646X cell lines (n = 12) and: EZH2WT cell lines (n = 4), EZH2Y646X cell lines treated with EPZ6438 (n = 12), and EZH2Y646X cell lines treated with GSK126 (n = 12). (a) Full density plot, (b) zoom of the bottom part. c, Comparison of TAD mRNA expression fold-changes (n = 2,024 TADs) obtained by comparing cell lines treated with GSK126 versus vehicle (DMSO) (X-axis) and cell lines treated with EPZ6438 versus vehicle (DMSO) (Y-axis). Pearson’s correlation is reported. Points are colored to reflect point density: cold colors indicate low density; warm colors indicate high density. d-f, TAD mRNA expression fold-changes obtained by comparing EZH2Y646X cell lines (n = 12) and EZH2WT cell lines (n = 4) (X-axis) versus EZH2Y646X cell lines (n = 12) and (d) EZH2Y646X cell lines treated with GSK126 (n = 12) or (e) EPZ6438 (n = 12) (Y-axis). (f) TAD mRNA expression fold-changes obtained by comparing OCI-Ly19-EZH2Y646F and OCI-Ly19-EZH2WT (X-axis) versus OCI-Ly19-EZH2Y646X cells treated with GSK126 (Y-axis, n = 3 cell lines per condition). The number of TADs within each quadrant is reported. Points are colored to reflect point density: cold colors indicate low density; warm colors indicate high density. g, Quantitative mRNA expression analysis of genes in the TAD6.139 (FOXO3, ARMC2 and SESN1) in the indicated cell lines treated with 2 μM GSK126 or 1 μM EPZ6438 or vehicle (DMSO) for 72 hours. Gene expression levels in cells treated with DMSO were used as reference. Mean values of two independent experiments are reported, bars indicate mean values, error bars correspond to 1 standard deviation. h, Quantitative expression analysis of genes in the TAD6.139 (FOXO3, ARMC2 and SESN1) in OCY-Ly19 EZH2WT or OCY-Ly19 EZH2Y646F treated with 2 μM GSK126 (n = 3 experiments) or 1 μM EPZ6438 (n = 2) or vehicle (DMSO) (n = 3) for 72 hours. Bars indicate mean values; error bars correspond to one standard deviation.
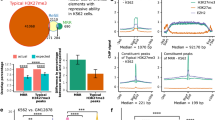
Supplementary Figure 7 Intra-TAD structural changes in EZH2WT and EZH2Y646X cells by interactome.
a, Hi-C contact map at 20kb resolution of the genomic region Chr.6 108.3-109.8Mb in Karpas422-EZH2Y646N (left) and Karpas422-EZH2Y646N treated with GSK126 for 72 hours (right). TAD boundaries are contoured in black and representative genes within the TADs are shown. b, Number of normalized paired-end reads between Chr.6 108,860-108,880 kb and Chr.6 109,380-109,400 kb in the indicated Hi-C cell lines and conditions. P-value derived by two-tailed Wilcoxon test comparing normalized read counts in EZH2Y646X (n = 4) cell lines and in EZH2WT or EZH2Y646X cell lines (n = 4 in total) treated with GSK126. c, Significant interactions in WSU-DLCL2-DMSO (top) or WSU-DLCL2-GSK126 (bottom) determined by HiC-DC. Bin-pairs within a 2Mb windows were tested (see Methods). d, Significantly different interactions (q-value < 0.1) between WSU-DLCL2-DMSO and WSU-DLCL2-GSK126 (n = 435 tested interactions). Empirical q-values determined as described in Methods. e, Most significantly different interactions between WSU-DLCL2-DMSO and WSU-DLCL2-GSK126. f, UMI-4C domainogram: mean number of contacts (% of the maximum) in the Chr.6 108.80-109.45kb region in Karpas422-DMSO (top) and Karpas422-GSK126 (bottom) using Primer 2. g, Normalized number of UMI-4C reads in Karpas422-DMSO and Karpas422-GSK126 obtained with Primer 2. Normalized read-counts are binned at 5kb and averaged within a sliding 5 bin-window. h, (Top) Most significant different interactions determined by HiC-DC between Karpas422-GSK126 than Karpas422-DMSO (see Methods). Blue: stronger in Karpas422-GSK126, Red: stronger in Karpas422-DMSO. (Bottom) ChIP-seq tracks for H3K4me3 (green), H3K27Ac (dark blue), H3K4me1 (light blue), and H3K27me3 (violet) in Karpas422 and OCI-Ly7 (n = 2 ChIP-seq experiments per cell line). i, (Left) Significant interactions in Karpas422-EZH2Y646N (top) and OCI-Ly19-EZH2WT (bottom) determined by HiC-DC. Bin-pairs within a 2Mb windows were tested (see Methods). (Right-top) Significantly different interactions (q-value < 0.1) between Karpas422-EZH2Y646N and OCI-Ly19-EZH2WT (n = 435 tested interactions). Empirical q-values determined as described in Methods. (Right-bottom) Most significantly different interactions between Karpas422-EZH2Y646N and OCI-Ly19-EZH2WT. j, Distribution of the number of significant different interactions between Karpas422-EZH2Y646N and OCI-Ly19-EZH2WT cells in in 100 sets of 72 randomly sampled neutral TADs and value observed in 72 inactive TADs (red line). Empirical p-value corresponds to fraction of random sets of neutral TADs with a number of significant interactions equal or greater to the one observed in inactive TADs. k, Venn diagram of the overlap between significant different interactions between Karpas422-EZH2Y646N and OCI-Ly19-EZH2WT. P-value derived by two-tailed Fisher’s exact test, OR: odds ratio.
Supplementary Figure 8 Intra-TAD structural changes in EZH2WT and EZH2Y646X cells by STORM.
a, Representative immunofluorescence images from one experiment with WSU-DLCL2 cells treated with 2 μM of GSK126 or DMSO for 72 hours and stained with H3K27me3 and DAPI. Scale bar = 20 μm. b, Graphical representation of data points retained in STORM analyses using a neighborhood radius equal to 40 nm (left) or 30 nm (right). c, Representative images overlaying wide field (gray-scale) and STORM acquisition of TAD6.139 in WSU-DLCL2 cells treated with 2 μM of GSK126 or DMSO for 72 hours in 4 biologically independent experiments. d, Representative images overlaying wide field and STORM acquisition of TAD6.139 in OCI-Ly19-EZH2Y646F (red) and OCI-Ly19-EZH2WT in 3 biologically independent experiments. e, Eccentricity of TAD6.139 with a (neighborhood radius = 30 nm) in WSU-DLCL2 treated with 2 μM of GSK126 or vehicle (DMSO) for 72 hours (n = 4 biologically independent experiments) and in OCI-Ly19 EZH2WT and OCI-Ly19 EZH2Y646 (n = 3 biologically independent experiments). Dots are means of multiple measurements of independent cells (Supplementary Table 7), error bars are +/-1 standard error. f, Representative images overlaying wide field and STORM acquisition of TAD1.54 in WSU-DLCL2 cells treated with 2 μM of GSK126 or DMSO for 72 hours (n = 2 biologically independent experiments). g, Eccentricity of TAD1.54 with neighborhood radius = 30 nm (left) and 40 nm (right) in 2 biologically independent experiments comparing WSU-DLCL2 treated with 2 μM of GSK126 or vehicle (DMSO) for 72 hours. Dots are means of multiple measurements of independent cells (Supplementary Table 7), error bars are +/- 1standard error.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Note
Supplementary Table 1
Hi-C in EZH2 wild-type and mutant cells
Supplementary Table 2
List of TADs with epigenetic and transcriptional annotations in cell lines, patient samples and cell treated with EZH2 inhibitors GSK126 and EPZ6438.
Supplementary Table 3
Gene-set enrichment analysis in inactive TADs
Supplementary Table 4
Percentage of cells expressing single or dual shRNA for the indicated genes in three independent experiments.
Supplementary Table 5
Interactome analysis in EZH2 wild-type and mutated cells and in cell treated with EZH2 inhibitor GSK126.
Supplementary Table 6
Number of mapped reads in UMI-4C seq
Supplementary Table 7
STORM measurements in WSU-DLCL2 cells treated with GSK126 or vehicle (control) for 72 h and in OCI-Ly19 EZH2WT and EZH2Y646F cells for the indicated TAD
Supplementary Table 8
List of primers used in this study
Rights and permissions
About this article
Cite this article
Donaldson-Collier, M.C., Sungalee, S., Zufferey, M. et al. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat Genet 51, 517–528 (2019). https://doi.org/10.1038/s41588-018-0338-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0338-y
This article is cited by
-
Metabolic heterogeneity in clear cell renal cell carcinoma revealed by single-cell RNA sequencing and spatial transcriptomics
Journal of Translational Medicine (2024)
-
EZH1/2 alteration as a potential biomarker for immune checkpoint inhibitors across multiple cancer types
Journal of Translational Medicine (2023)
-
Widespread contribution of transposable elements to the rewiring of mammalian 3D genomes
Nature Communications (2023)
-
Loss of cohesin regulator PDS5A reveals repressive role of Polycomb loops
Nature Communications (2023)
-
Whole-genome doubling drives oncogenic loss of chromatin segregation
Nature (2023)