Abstract
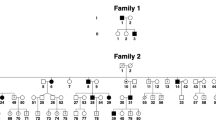
Late-onset ataxia is common, often idiopathic, and can result from cerebellar, proprioceptive, or vestibular impairment; when in combination, it is also termed cerebellar ataxia, neuropathy, vestibular areflexia syndrome (CANVAS). We used non-parametric linkage analysis and genome sequencing to identify a biallelic intronic AAGGG repeat expansion in the replication factor C subunit 1 (RFC1) gene as the cause of familial CANVAS and a frequent cause of late-onset ataxia, particularly if sensory neuronopathy and bilateral vestibular areflexia coexist. The expansion, which occurs in the poly(A) tail of an AluSx3 element and differs in both size and nucleotide sequence from the reference (AAAAG)11 allele, does not affect RFC1 expression in patient peripheral and brain tissue, suggesting no overt loss of function. These data, along with an expansion carrier frequency of 0.7% in Europeans, implies that biallelic AAGGG expansion in RFC1 is a frequent cause of late-onset ataxia.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The genotyping microarray and sequence data obtained by WGS and RNA-seq are available upon request from the corresponding authors (A.C., H.H.). They are not publicly available because some of the study participants did not give their full consent to release data publicly. Since WGS data are protected by the Personal Information Protection Law, availability of these data is under the regulation of the institutional review board. The data obtained from RNA-seq have been deposited on the NCBI Sequence Read Archive under accession no. SRP186868.
Change history
26 April 2019
In the version of this article initially published, the name of author Wai Yan Yau was misspelled. The error has been corrected in the HTML and PDF versions of the article.
References
Harding, A. E. “Idiopathic” late onset cerebellar ataxia. A clinical and genetic study of 36 cases. J. Neurol. Sci. 51, 259–271 (1981).
Muzaimi, M. B. et al. Population based study of late onset cerebellar ataxia in south east Wales. J. Neurol. Neurosurg. Psychiatry 75, 1129–1134 (2004).
Sghirlanzoni, A., Pareyson, D. & Lauria, G. Sensory neuron diseases. Lancet Neurol. 4, 349–361 (2005).
Strupp, M., Feil, K., Dieterich, M. & Brandt, T. Bilateral vestibulopathy. Handb. Clin. Neurol. 137, 235–240 (2016).
Abele, M. et al. The aetiology of sporadic adult-onset ataxia. Brain 125(Pt. 5), 961–968 (2002).
Kirchner, H. et al. Clinical, electrophysiological, and MRI findings in patients with cerebellar ataxia and a bilaterally pathological head-impulse test. Ann. N. Y. Acad. Sci. 1233, 127–138 (2011).
Migliaccio, A. A., Halmagyi, G. M., McGarvie, L. A. & Cremer, P. D. Cerebellar ataxia with bilateral vestibulopathy: description of a syndrome and its characteristic clinical sign. Brain 127(Pt. 2), 280–293 (2004).
Szmulewicz, D. J. et al. Proposed diagnostic criteria for cerebellar ataxia with neuropathy and vestibular areflexia syndrome (CANVAS). Neurol. Clin. Pract. 6, 61–68 (2016).
Wu, T. Y. et al. Autonomic dysfunction is a major feature of cerebellar ataxia, neuropathy, vestibular areflexia ‘CANVAS’ syndrome. Brain 137(Pt. 10), 2649–2656 (2014).
Szmulewicz, D. J., Merchant, S. N. & Halmagyi, G. M. Cerebellar ataxia with neuropathy and bilateral vestibular areflexia syndrome: a histopathologic case report. Otol. Neurotol. 32, e63–e65 (2011).
Szmulewicz, D. J. et al. Dorsal root ganglionopathy is responsible for the sensory impairment in CANVAS. Neurology 82, 1410–1415 (2014).
Cazzato, D., Bella, E. D., Dacci, P., Mariotti, C. & Lauria, G. Cerebellar ataxia, neuropathy, and vestibular areflexia syndrome: a slowly progressive disorder with stereotypical presentation. J. Neurol. 263, 245–249 (2016).
Rust, H. et al. VEMPs in a patient with cerebellar ataxia, neuropathy and vestibular areflexia (CANVAS). J. Neurol. Sci. 378, 9–11 (2017).
Pelosi, L. et al. Peripheral nerve ultrasound in cerebellar ataxia neuropathy vestibular areflexia syndrome (CANVAS). Muscle Nerve. 56, 160–162 (2017).
Pelosi, L. et al. Peripheral nerves are pathologically small in cerebellar ataxia neuropathy vestibular areflexia syndrome: a controlled ultrasound study. Eur. J. Neurol. 25, 659–665 (2018).
Taki, M. et al. Cerebellar ataxia with neuropathy and vestibular areflexia syndrome (CANVAS). Auris Nasus Larynx 45, 866–870 (2018).
Infante, J. et al. Cerebellar ataxia, neuropathy, vestibular areflexia syndrome (CANVAS) with chronic cough and preserved muscle stretch reflexes: evidence for selective sparing of afferent Ia fibres. J. Neurol. 265, 1454–1462 (2018).
Hommelsheim, C. M., Frantzeskakis, L., Huang, M. & Ülker, B. PCR amplification of repetitive DNA: a limitation to genome editing technologies and many other applications. Sci. Rep. 4, 5052 (2014).
Campuzano, V. et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 (1996).
Orr, H. T. et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 4, 221–226 (1993).
Pulst, S. M. et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 14, 269–276 (1996).
Kawaguchi, Y. et al. CAG expansions in a novel gene for Machado–Joseph disease at chromosome 14q32.1. Nat Genet. 8, 221–228 (1994).
Lupski, J. R. et al. DNA duplication associated with Charcot–Marie–Tooth disease type 1A. Cell 66, 219–232 (1991).
Hayasaka, K. et al. Charcot–Marie–Tooth neuropathy type 1B is associated with mutations of the myelin P0 gene. Nat. Genet. 5, 31–34 (1993).
Bergoffen, J. et al. Connexin mutations in X-linked Charcot–Marie–Tooth disease. Science 262, 2039–2042 (1993).
Züchner, S. et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot–Marie–Tooth neuropathy type 2A. Nat. Genet. 36, 449–451 (2004).
Fridman, V. et al. CMT subtypes and disease burden in patients enrolled in the Inherited Neuropathies Consortium natural history study: a cross-sectional analysis. J. Neurol. Neurosurg. Psychiatry 86, 873–878 (2015).
Murphy, S. M. et al. Charcot–Marie–Tooth disease: frequency of genetic subtypes and guidelines for genetic testing. J. Neurol. Neurosurg. Psychiatry 83, 706–710 (2012).
Seixas, A. I. et al. A pentanucleotide ATTTC repeat insertion in the non-coding region of DAB1, mapping to SCA37, causes spinocerebellar ataxia. Am. J. Hum. Genet. 101, 87–103 (2017).
Ishiura, H. et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat. Genet. 50, 581–590 (2018).
Deininger, P. Alu elements: know the SINEs. Genome Biol. 12, 236 (2011).
Haeusler, A. R., Donnelly, C. J. & Rothstein, J. D. The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat. Rev. Neurosci. 17, 383–395 (2016).
Dürr, A. et al. Clinical and genetic abnormalities in patients with Friedreich’s ataxia. N. Engl. J. Med. 335, 1169–1175 (1996).
Lazaropoulos, M. et al. Frataxin levels in peripheral tissue in Friedreich ataxia. Ann. Clin. Transl. Neurol. 2, 831–842 (2015).
Paulson, H. Repeat expansion diseases. Handb. Clin. Neurol. 147, 105–123 (2018).
Majka, J. & Burgers, P. M. J. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 78, 227–260 (2004).
Tomida, J. et al. DNA damage-induced ubiquitylation of RFC2 subunit of replication factor C complex. J. Biol. Chem. 283, 9071–9079 (2008).
Overmeer, R. M. et al. Replication factor C recruits DNA polymerase delta to sites of nucleotide excision repair but is not required for PCNA recruitment. Mol. Cell. Biol. 30, 4828–4839 (2010).
McKinnon, P. J. Maintaining genome stability in the nervous system. Nat. Neurosci. 16, 1523–1529 (2013).
Higuchi, Y. et al. Mutations in MME cause an autosomal-recessive Charcot–Marie–Tooth disease type 2. Ann. Neurol. 79, 659–672 (2016).
La Spada, A. R. & Taylor, J. P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 11, 247–258 (2010).
Sznajder, Ł. J. et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl Acad. Sci. USA 115, 4234–4239 (2018).
Gebus, O. et al. Deciphering the causes of sporadic late-onset cerebellar ataxias: a prospective study with implications for diagnostic work. J. Neurol. 264, 1118–1126 (2017).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011).
Aneichyk, T. et al. Dissecting the causal mechanism of X-linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell 172, 897–909.e21 (2018).
Manole, A. et al. Clinical, pathological and functional characterization of riboflavin-responsive neuropathy. Brain 140, 2820–2837 (2017).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Abecasis, G. R., Cherny, S. S., Cookson, W. O. & Cardon, L. R. Merlin: rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Layer, R. M., Chiang, C., Quinlan, A. R. & Hall, I. M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15, R84 (2014).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Weis, J., Brandner, S., Lammens, M., Sommer, C. & Vallat, J.-M. Processing of nerve biopsies: a practical guide for neuropathologists. Clin. Neuropathol. 31, 7–23 (2012).
Dubowitz, V, Sewry, C. A. & Oldfors, A. Muscle Biopsy: a Practical Approach (Elsevier, 2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Anders, S., Reyes, A. & Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 (2012).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Reimand, J. et al. g:Profiler: a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44, W83–W89 (2016).
Paz, I., Kosti, I., Ares, M. Jr., Cline, M. & Mandel-Gutfreund, Y. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 42, W361–W367 (2014).
Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A. & Eddy, S. R. Rfam: an RNA family database. Nucleic Acids Res. 31, 439–441 (2003).
Podhorecka, M., Skladanowski, A. & Bozko, P. H2AX phosphorylation: its role in DNA damage response and cancer therapy. J. Nucleic Acids 2010, 920161 (2010).
Sharma, A., Singh, K. & Almasan, A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 920, 613–626 (2012).
Acknowledgements
A.C. is funded by the Inherited Neuropathies Consortium (INC), which is part of the National Institutes of Health Rare Diseases Clinical Research Network (RDCRN) (grant no. U54NS065712) and the Wellcome Trust (grant no. 204841/Z/16/Z). A.M.R. is funded by a Wellcome Trust Postdoctoral Fellowship for Clinicians (no. 110043/Z/15/Z). H.H. is supported by the Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Muscular Dystrophy UK, Muscular Dystrophy Association, Higher Education Commission of Pakistan and Wellcome Trust (Synaptopathies Strategic Award, 165908). M.M.R. is grateful to the Medical Research Council (MRC), MRC Centre grant (G0601943) and to the National Institutes of Neurological Diseases and Stroke and Office of Rare Diseases (U54NS065712) for their support. The INC (grant no. U54NS065712) is part of the National Center for Advancing Translational Sciences (NCATS) RDCRN. The RDCRN is an initiative of the NCATS Office of Rare Diseases Research and is funded through a collaboration between NCATS and the National Institute of Neurological Disorders and Stroke. S.Z. thanks the National Institutes of Health (NIH; grant no. 4R01NS075764) for its support. This research was also supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre (176718). Neuromuscular and brain tissue samples were obtained from University College London Hospitals NHS Foundation Trust as part of the UK Brain Archive Information Network (BRAIN UK), which is funded by the Medical Research Council and Brain Tumour Research and the NIH-funded NeuroBioBank. We also thank F. Launchbury from the UCL IQPath laboratory for her technical assistance in histology slide preparation. We would also like to thank Muscular Dystrophy UK and Muscular Dystrophy Association USA (award 171011). P.F. is funded by an MRC/MNDA CSF (MR/M008606/1).
Author information
Authors and Affiliations
Contributions
A.C. designed the study, collected the clinical data, performed the genetic analysis that led to the discovery of the AAGGG repeat expansions, analyzed the data, and drafted the manuscript together with contributions from J.V., R.Simone, R.Sullivan, and J.H. R.Simone, N.S.A., E.T., E.B., A.R., W.Y.Y., and M.I. performed the investigation on RFC1 expression. J.V. performed the computational genetic analysis. R.Sullivan and H.T. collected and analyzed the genetic data in healthy controls. P.J.T., W.J.M., A.B., G.D., I.C., M.V., D.K., V.S., S.E., N.W.W., and A.M.R. contributed with the collection of clinical data and patient samples. J.H., P.S., and P.F. performed the RNA-seq analysis. Z.J. performed the pathological investigation. R.Simone, A.M.R., P.F., and J.P. contributed to the design of the study. S.Z. contributed to the design of the study and analyzed the data. H.H. and M.M.R. designed the study, collected the patient clinical data and biological samples and analyzed the data. All authors revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Homozygosity mapping in a consanguineous CANVAS family.
(a,b) Homozygosity mapping plot (a) and genomic positions (b) defining the homozygous regions shared by Fam7-1 and Fam7-2. The blue arrow in a points to a ~ 12-Mb homozygous region on chromosome 4 (bold highlighted in b), which encompass the AAGGG repeat expansion locus.
Supplementary Figure 2 Southern blots.
Southern blotting of genomic DNA from 18 patients from 11 families and 16 sporadic cases. Patients show two discrete or overlapping bands of 7 to 15 kb. Unaffected siblings, indicated by “ * ”, carry an expanded allele and one allele in the normal range. In controls (CTRL), one 5-kb band corresponding to the expected size for reference allele (AAAAG)11 is usually observed, but bands of increased size can also be seen. In some blots, an unspecific band at 2 kb is also observed. Ladder used are DIG-labelled DNA Molecular Weight Marker II (Roche) (LADDER II, left) containing 8 fragments with the following base pair lengths: 125 (not shown), 564, 2,027, 2,322, 4,361, 6,557, 9,416, and 23,130 bp and DIG-labelled DNA Molecular Weight Marker III (Roche) (LADDER III, right) containing 13 fragments with the following base pair lengths: 125 (not shown), 564, 831, 947, 1,375, 1,584, 1,904, 2,027, 3,530, 4,268, 4,973, 5,148, and 21,226 bp. N.I., sample not included in this study; rep, repeated sample.
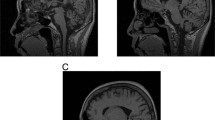
Supplementary Figure 3 Morphological findings in the brain from patient with CANVAS.
(a) Histological examination reveals some age-related changes in the medial temporal lobe with frequent diffuse parenchymal amyloid-β deposits (a) in the neocortex, subiculum, CA1 and caudate nucleus (Thal phase 3). Rare leptomeningeal amyloid angiopathy (not shown) is also seen in the cerebral hemisphere. Neurofibrillary tangle tau pathology (d) is restricted to the medial temporal lobe with the extent corresponding to Braak and Braak stage IV. There are no α-synuclein (g) or TDP43 (h) positive pathological inclusions in the medial temporal lobe. (b) In the brainstem, the locus coeruleus (b) and pontine base nuclei, and white matter tracts show no apparent neuronal loss. In the midbrain, the substantia nigra shows no significant reduction of pigmented neurones (e). In the inferior olivary nucleus, however, there is widespread depletion of neurones and marked chronic gliosis (i). (c) In the cerebellar hemisphere and cerebellar vermis, haematoxylin and eosin stained section shows prominent cortical atrophy with more prominent atrophy seen in the superior cerebellar vermis (c and f, blue square) when compared with the inferior cerebellar vermis (c and j, red square). There are no p62 positive pathological inclusions (f and j) in the cerebellum. Scale bar: 3 mm in a and d; 200 µm in g and e; 40 µm in h; 100 µm in b, i, f and j; 35 mm in c. Stainings were carried out once on patients’ samples with appropriate controls according to standard practice and histopathology procedures in an ISO15189 accredited laboratory.
Supplementary Figure 4 Morphological findings in the nerve and muscle biopsy from a representative patient with CANVAS.
Sural nerve biopsy (a, c, and e). a and c, semi-thin resin section stained with methylene blue azure-basic fuchsin; e, immunostaining for neurofilament with SMI31. Quadriceps skeletal muscle biopsy (b, d and f). b, stained with haematoxylin and eosin; d and f, histochemical staining with ATP 4.2 (d) and ATP9.4 (f). The nerve biopsy demonstrates marked loss of large and small myelinated fibres (a, c, and e), blue arrowheads) with no active axonal degeneration or any signs of regeneration. The unmyelinated fibres (e) are comparably better preserved. The skeletal muscle biopsy reveals angulated fibres and particularly small atrophic fibres (b, yellow arrowhead). There is grouping of type 1 and type 2 fibres (d red arrows and f), in keeping with chronic denervation with reinnervation. Scale bar: 100 µm in a, d and f; 50 µm in c, e and b. Stainings were carried out once on patients’ samples with appropriate controls according to standard practice and histopathology procedures in an ISO15189 accredited laboratory.
Supplementary Figure 5 RNA fluorescence in situ hybridization (FISH).
(a-e) No endogenous RNA foci were detected using oligonucleotides (AAGGG)5 (a, b, c, d, e) or (TTCCC)5 (f, g, h, I, j) probes targeting transcripts containing sense (TTCCC) or antisense (GGGAA) repeated units in unaffected individuals (a, f), pathological controls (b, g), or CANVAS patient (c, h). As a technical control, SH-SY5Y cells were transfected with plasmids expressing mutant sense (TTCCC)94 (d), mutant anti-sense (AAGGG)54 (i), wild-type sense (TTTTC)11 (e) or wild-type anti-sense (AAAAG)11 (j) and analyzed 24 h after transfection by RNA FISH. RNA foci were detected in d and i (red). Nuclei were stained with DAPI (blue). Scale bar: 20 µm in a, b, c, f, g, h; 10 µm in d, e, i, j. Experiments were repeated independently twice with similar results.
Supplementary Figure 6 RFC1 intron 2 retention in multiple tissue types in CANVAS.
Expression levels of intron 2 in RFC1 pre-mRNA as measured by qRT-PCR using primers cF1-iR1 in control (n = 3) and CANVAS (n = 2) lymphoblasts; control (n = 3), Friedreich’s ataxia (n = 3) and CANVAS (n = 1) cerebellum and frontal cortex; control (n = 5) and CANVAS muscles (n = 7). Bar graphs show mean ± s.d. and data distribution (black dots). Two-tailed t-test was performed to compare the expression level of intron 2 in RFC1 pre-mRNA in patients versus healthy or disease controls. All experiments were repeated independently twice with similar results. CANVAS, cerebellar ataxia, neuropathy, vestibular areflexia syndrome; CBM, cerebellum; Ctrl, control; FCX, frontal cortex; FRDA, Friedreich’s ataxia; IR, intron 2 retention; LBLs, lymphoblasts.
Supplementary Figure 7 Western blots for RFC1-encoded protein expression.
(a,b) Uncropped gels showing RFC1-encoded protein levels as measured by Western blotting using the polyclonal antibody (GTX129291) and normalized to β-actin in (a) control (n = 5, lanes 1-5) and CANVAS (n = 5, lanes 6-10) fibroblasts, control (n = 3, lanes 12-14) and CANVAS (n = 4, lanes 15-18) lymphoblasts and (b) CANVAS (n = 1, lane 2), control (n = 3, lanes 3-5), Friedreich’s ataxia (n = 3, lanes 6-8) vermis and CANVAS (n = 1, lane 9), control (n = 3, lanes 10-12) and Friedreich’s ataxia (n = 3, lanes 13-15) frontal cortex. Green arrow indicates the expected 128 kDa band corresponding to RFC1-encoded protein, and the red arrow a 45 kDa band of β-actin. Ladder in lanes 11 (a) and 1 (b) is SeeBlue Plus2 (Thermofisher).
Supplementary Figure 8 Response to DNA damage in CANVAS.
(a) Cell viability after DNA damage inducing treatments of CANVAS (n = 5) and control (n = 5) fibroblasts. (b) Quantification of γH2AX protein expression levels normalized to actin and plotted relative to controls. (c) Representative western blot of cells treated with different DNA damage inducing agents. Bar graphs show mean ± s.d. and data distribution (dots). All experiments were repeated independently twice with similar results.
Supplementary Information
Supplementary Information
Supplementary Figures 1–8 and Supplementary Tables 1–3
Supplementary Data
Supplementary Data
Rights and permissions
About this article
Cite this article
Cortese, A., Simone, R., Sullivan, R. et al. Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat Genet 51, 649–658 (2019). https://doi.org/10.1038/s41588-019-0372-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0372-4
This article is cited by
-
Origin and evolution of pentanucleotide repeat expansions at the familial cortical myoclonic tremor with epilepsy type1 - SAMD12 locus
European Journal of Human Genetics (2024)
-
Clinical and functional characteristics, possible causes, and impact of chronic cough in patients with cerebellar ataxia, neuropathy, and bilateral vestibular areflexia syndrome (CANVAS)
Journal of Neurology (2024)
-
Brain 18F-FDG PET findings and sequential vestibular testing in SCA27B: a case report
Journal of Neurology (2024)
-
Clinical and genetic analyses of a Swedish patient series diagnosed with ataxia
Journal of Neurology (2024)
-
Comprehensive Analysis of a Japanese Pedigree with Biallelic ACAGG Expansions in RFC1 Manifesting Motor Neuronopathy with Painful Muscle Cramps
The Cerebellum (2024)