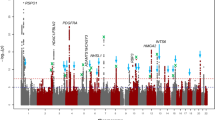
Abstract
Refractive errors, in particular myopia, are a leading cause of morbidity and disability worldwide. Genetic investigation can improve understanding of the molecular mechanisms that underlie abnormal eye development and impaired vision. We conducted a meta-analysis of genome-wide association studies (GWAS) that involved 542,934 European participants and identified 336 novel genetic loci associated with refractive error. Collectively, all associated genetic variants explain 18.4% of heritability and improve the accuracy of myopia prediction (area under the curve (AUC) = 0.75). Our results suggest that refractive error is genetically heterogeneous, driven by genes that participate in the development of every anatomical component of the eye. In addition, our analyses suggest that genetic factors controlling circadian rhythm and pigmentation are also involved in the development of myopia and refractive error. These results may enable the prediction of refractive error and the development of personalized myopia prevention strategies in the future.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Summary statistics from the cohorts that participated in the meta-analysis can be downloaded from ftp://twinr-ftp.kcl.ac.uk/Refractive_Error_MetaAnalysis_2020 and public repositories such as the GWAS Catalog (https://www.ebi.ac.uk/gwas/downloads/summary-statistics). These freely downloadable summary statistics are calculated using all cohorts described in this manuscript, except for the 23andMe participants. This is due to a non-negotiable clause in the 23andMe data transfer agreement, intended to protect the privacy of the 23andMe research participants.
To fully recreate our meta-analytic results, all bona fide researchers can obtain the 23andMe summary statistics by emailing 23andMe (dataset-request@23andme.com) and subsequently meta-analyzing them along with the freely accessible summary statistics for all the other cohorts.
References
Vos, T. et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1211–1259 (2017).
The Global Burden of Disease: 2004 Update (World Health Organization, 2008).
Williams, K. M. et al. Increasing prevalence of myopia in Europe and the impact of education. Ophthalmology 122, 1489–1497 (2015).
Sanfilippo, P. G., Hewitt, A. W., Hammond, C. J. & Mackey, D. A. The heritability of ocular traits. Surv. Ophthalmol. 55, 561–583 (2010).
Kiefer, A. K. et al. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 9, e1003299 (2013).
Verhoeven, V. J. et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet. 45, 314–318 (2013).
Tedja, M. S. et al. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat. Genet. 50, 834–848 (2018).
Cumberland, P. M. et al. Frequency and distribution of refractive error in adult life: methodology and findings of the UK Biobank study. PLoS One 10, e0139780 (2015).
Kvale, M. N. et al. Genotyping informatics and quality control for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics 200, 1051–1060 (2015).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Dudbridge, F. & Gusnanto, A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 (2008).
Pe’er, I., Yelensky, R., Altshuler, D. & Daly, M. J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32, 381–385 (2008).
Wood, A. R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Oliveira, J. et al. LAMA2 gene analysis in a cohort of 26 congenital muscular dystrophy patients. Clin. Genet. 74, 502–512 (2008).
Colognato, H. et al. Identification of dystroglycan as a second laminin receptor in oligodendrocytes, with a role in myelination. Development 134, 1723–1736 (2007).
Burkin, D. J. & Kaufman, S. J. The α7β1 integrin in muscle development and disease. Cell Tissue Res. 296, 183–190 (1999).
Ervasti, J. M. & Campbell, K. P. Dystrophin-associated glycoproteins: their possible roles in the pathogenesis of Duchenne muscular dystrophy. Mol. Cell Biol. Hum. Dis. Ser. 3, 139–166 (1993).
Mayer, U. et al. Absence of integrin α7 causes a novel form of muscular dystrophy. Nat. Genet. 17, 318–323 (1997).
Jean, D., Ewan, K. & Gruss, P. Molecular regulators involved in vertebrate eye development. Mech. Dev. 76, 3–18 (1998).
Hammond, C. J., Andrew, T., Mak, Y. T. & Spector, T. D. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am. J. Hum. Genet. 75, 294–304 (2004).
Ali, M. et al. Null mutations in LTBP2 cause primary congenital glaucoma. Am. J. Hum. Genet. 84, 664–671 (2009).
Clark, A. M. et al. Negative regulation of Vsx1 by its paralog Chx10/Vsx2 is conserved in the vertebrate retina. Brain Res. 1192, 99–113 (2008).
Heon, E. et al. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum. Mol. Genet. 11, 1029–1036 (2002).
Hysi, P. G. et al. Common mechanisms underlying refractive error identified in functional analysis of gene lists from genome-wide association study results in 2 European British cohorts. JAMA Ophthalmol. 132, 50–56 (2014).
Ma, M. et al. Wnt signaling in form deprivation myopia of the mice retina. PLoS One 9, e91086 (2014).
Miyake, M. et al. Identification of myopia-associated WNT7B polymorphisms provides insights into the mechanism underlying the development of myopia. Nat. Commun. 6, 6689 (2015).
Cuellar-Partida, G. et al. WNT10A exonic variant increases the risk of keratoconus by decreasing corneal thickness. Hum. Mol. Genet. 24, 5060–5068 (2015).
Stone, R. A. et al. Image defocus and altered retinal gene expression in chick: clues to the pathogenesis of ametropia. Invest. Ophthalmol. Vis. Sci. 52, 5765–5777 (2011).
Zhou, H., Yoshioka, T. & Nathans, J. Retina-derived POU-domain factor-1: a complex POU-domain gene implicated in the development of retinal ganglion and amacrine cells. J. Neurosci. 16, 2261–2274 (1996).
Hysi, P. G. et al. Genome-wide association meta-analysis of individuals of European ancestry identifies new loci explaining a substantial fraction of hair color variation and heritability. Nat. Genet. 50, 652–656 (2018).
Fabian-Jessing, B. K. et al. Ocular albinism with infertility and late-onset sensorineural hearing loss. Am. J. Med. Genet. A 176, 1587–1593 (2018).
Thorleifsson, G. et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science 317, 1397–1400 (2007).
Rivera, A. et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum. Mol. Genet. 14, 3227–3236 (2005).
Morimura, H., Saindelle-Ribeaudeau, F., Berson, E. L. & Dryja, T. P. Mutations in RGR, encoding a light-sensitive opsin homologue, in patients with retinitis pigmentosa. Nat. Genet. 23, 393–394 (1999).
Robinson, P. N. et al. Mutations of FBN1 and genotype-phenotype correlations in Marfan syndrome and related fibrillinopathies. Hum. Mutat. 20, 153–161 (2002).
Rohde, K., Moller, M. & Rath, M. F. Homeobox genes and melatonin synthesis: regulatory roles of the cone-rod homeobox transcription factor in the rodent pineal gland. Biomed. Res. Int. 2014, 946075 (2014).
Chakraborty, R. et al. Circadian rhythms, refractive development, and myopia. Ophthalmic Physiol. Opt. 38, 217–245 (2018).
Carnes, M. U., Allingham, R. R., Ashley-Koch, A. & Hauser, M. A. Transcriptome analysis of adult and fetal trabecular meshwork, cornea, and ciliary body tissues by RNA sequencing. Exp. Eye Res. 167, 91–99 (2018).
Ratnapriya, R. et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 51, 606–610 (2019).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Westra, H. J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Qi, T. et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 9, 2282 (2018).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Benyamin, B. et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol. Psychiatry 19, 253–258 (2014).
Mountjoy, E. et al. Education and myopia: assessing the direction of causality by mendelian randomisation. BMJ 361, k2022 (2018).
Pozarickij, A. et al. Quantile regression analysis reveals widespread evidence for gene-environment or gene-gene interactions in myopia development. Commun. Biol. 2, 167 (2019).
Audo, I. et al. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 85, 720–729 (2009).
Khawaja, A. P. et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet. 50, 778–782 (2018).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Zhang, Y., Qi, G., Park, J. H. & Chatterjee, N. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat. Genet. 50, 1318–1326 (2018).
Zadnik, K. et al. Normal eye growth in emmetropic schoolchildren. Optom. Vis. Sci. 81, 819–828 (2004).
Li, Z. et al. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am. J. Hum. Genet. 85, 711–719 (2009).
Chow, R. L. et al. Vsx1, a rapidly evolving paired-like homeobox gene expressed in cone bipolar cells. Mech. Dev. 109, 315–322 (2001).
Struck, M. C. Albinism: update on ocular features. Curr. Ophthalmol. Rep. 3, 232–237 (2015).
Mohammad, S. et al. Characterization of abnormal optic nerve head morphology in albinism using optical coherence tomography. Invest. Ophthalmol. Vis. Sci. 56, 4611–4618 (2015).
Yahalom, C. et al. Refractive profile in oculocutaneous albinism and its correlation with final visual outcome. Br. J. Ophthalmol. 96, 537–539 (2012).
Lopez, V. M., Decatur, C. L., Stamer, W. D., Lynch, R. M. & McKay, B. S. L-DOPA is an endogenous ligand for OA1. PLoS Biol. 6, e236 (2008).
Karouta, C. & Ashby, R. S. Correlation between light levels and the development of deprivation myopia. Invest. Ophthalmol. Vis. Sci. 56, 299–309 (2014).
Wu, P. C., Tsai, C. L., Wu, H. L., Yang, Y. H. & Kuo, H. K. Outdoor activity during class recess reduces myopia onset and progression in school children. Ophthalmology 120, 1080–1085 (2013).
Troilo, D., Gottlieb, M. D. & Wallman, J. Visual deprivation causes myopia in chicks with optic nerve section. Curr. Eye Res. 6, 993–999 (1987).
de Leeuw, C. A., Neale, B. M., Heskes, T. & Posthuma, D. The statistical properties of gene-set analysis. Nat. Rev. Genet. 17, 353–364 (2016).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Delaneau, O., Marchini, J. & Zagury, J. F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2011).
Loh, P.-R., Kichaev, G., Gazal, S., Schoech, A. P. & Price, A. L. Mixed-model association for biobank-scale datasets. Nat. Genet. 50, 906–908 (2018).
Fuchsberger, C., Abecasis, G. R. & Hinds, D. A. minimac2: faster genotype imputation. Bioinformatics 31, 782–784 (2015).
Banda, Y. et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics 200, 1285–1295 (2015).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Riboli, E. & Kaaks, R. The EPIC Project: rationale and study design. European prospective investigation into cancer and nutrition. Int. J. Epidemiol. 26 (Suppl 1), S6–S14 (1997).
Hayat, S. A. et al. Cohort profile: a prospective cohort study of objective physical and cognitive capability and visual health in an ageing population of men and women in Norfolk (EPIC-Norfolk 3). Int. J. Epidemiol. 43, 1063–1072 (2014).
Delaneau, O., Marchini, J. & The 1000 Genomes Project Consortium. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 5, 3934 (2014).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Segre, A. V. et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010).
Pybus, M. et al. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42, D903–D909 (2014).
Acknowledgements
P.T.K. and P.J.F oversaw the UK Biobank eye data acquisition with support from the National Institute for Health Research (NIHR), Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology. The UK Biobank Eye and Vision Consortium was supported by grants from UK NIHR (BRC3_026), Moorfields Eye Charity (ST 15 11 E), Fight for Sight (1507/1508), The Macular Society, The International Glaucoma Association (IGA, Ashford UK) and Alcon Research Institute. V.V. is supported by a core UK Medical Research Council (MRC) grant MC_UU_00007/10. 23andMe thanks research participants and employees of 23andMe for making this work possible (a list of contributing staff is available in the Supplementary Note). Genotyping of the GERA cohort was funded by the US National Institute on Aging, the National Institute of Mental Health and the National Institute of Health Common Fund (RC2 AG036607); data analyses were funded by the National Eye Institute (NEI R01 EY027004, E.J.) and the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK116738, E.J.). The CREAM GWAS meta-analysis was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (grant 648268 to C.C.W.K), the Netherlands Organisation for Scientific Research (NWO, 91815655 to C.C.W.K) and the National Eye Institute (R01EY020483). V.J.M.V. acknowledges funding from the Netherlands Organisation for Scientific Research (NWO, grant 91617076). S.M. acknowledges support from the National Health and Medical Research Council (NHMRC) of Australia (grants 1150144, 1116360, 1154543, 1121979). EPIC-Norfolk infrastructure and core functions are supported by the MRC (G1000143) and Cancer Research UK (C864/A14136). Genotyping was funded by the MRC (MC_PC_13048). A.K.P. is supported by a Moorfields Eye Charity grant. P.J.F. received support from the Richard Desmond Charitable Trust, the National Institute for Health Research to Moorfields Eye Hospital and the Biomedical Research Centre for Ophthalmology. RW and PGH were supported by the National Eye Institute of the National Institutes of Health under award number R21EY029309. M.J.S. is a recipient of a Fight for Sight PhD studentship. K.P. is a recipient of a Fight for Sight PhD studentship. P.G.H. is the recipient of a FfS ECI fellowship. P.G.H. and C.J.H. acknowledge the TFC Frost Charitable Trust Support for the KCL Department of Ophthalmology. Statistical analyses were run in King’s College London on the Rosalind HPC LINUX Clusters and cloud servers. The UK Biobank data were accessed as part of the UK Biobank projects 669 and 17615. J.S.R. is supported in part by the NIHR Biomedical Research Centres at Moorfields Eye Hospital and the UCL Institute of Ophthalmology, and at the UCL Institute of Child Health and Great Ormond Street Hospital, and is an NIHR Senior Investigator. P.M.C. was funded by the Ulverscroft Foundation. O.A.M is supported by Wellcome Trust grant 206619_Z_17_Z and the NIHR Biomedical Research Centre at Moorfields Eye Hospital and the UCL Institute of Ophthalmology.
Author information
Authors and Affiliations
Consortia
Contributions
P.G.H., J.S.R., E.J. and C.J.H. conceived and designed the study. P.T.K., P.J.F. and J.S.R. contributed to the collection of data. P.G.H., H.C., A.P.K., R.W., M.S.T., J.Y., K.K.T., P.M.C., V.V., J.A.G and E.J. performed statistical analysis. A.P.K., M.J.S., K.P., K.K.T., AS and J.A.G. performed post-GWAS follow-up analyses. P.G.H., H.C., A.P.K., R.W., J.S.R., E.J. and C.J.H. wrote the manuscript with help from O.A.M., P.M.C., R.B.M., V.J.M.V., A.S., R.A.S., N.W., A.W.H., D.A.M., C.C.W.K., S.M., P.T.K., P.J.F. and J.A.G. who helped with the interpretation of the results.
Corresponding author
Ethics declarations
Competing interests
23andMe is a consumer genomics company.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Principal components plots of the subjects in the main participating cohorts.
a) UK Biobank (including the 102,117 subjects with direct refraction measurement and the imputed 108,956 likely myopes to 70,941 likely non-myopes, for a total of 179,897 subjects), b) Genetic Epidemiology Research on Adult Health and Aging (GERA, N = 34,998), c) 23andMe (106,086 cases and 85,757 controls, or 191,843 subjects in total).
Extended Data Fig. 2 Correlation of effect sizes between the discovery cohort meta-analysis.
Effect sizes are from two analyses, discovery (UK Biobank analysis on spherical equivalent + GERA, spherical equivalent + 23andMe, self-reported myopia cases and controls + UK Biobank inferred myopia cases and controls, for a total of N = 508,855 subjects) and the replication from the non-British CREAM Consortium participants (N = 34,079), used as replication. The z-scores for the discovery are on the y-axis and those from the CREAM cohort in the x-axis.
Extended Data Fig. 3 Distribution of the base-pair length (red) of the 449 regions associated in the meta-analysis of all available cohorts (from Supplementary Table 3), alongside the distribution of number of SNPs (blue) for each region.
Numbers in each of the axes in the figure are differentially colored to match the density curve they correspond to: red for the length of the region and blue for the number of SNPs.
Extended Data Fig. 4 Expression of genes located in the associated loci (from Supplementary Table 3) along the x-axis, across several human body tissues (y-axis).
The colors represent the centile ranking of the expression level of the gene in the tissue of interest. The hotter colors represent higher ranking of the gene expression and the colder colors low expression. Both genes and tissues are clustered in accordance with their pattern similarity. The symbol of all the genes could not be visualized and therefore are removed for the sake of clarity. Eye tissues, whether fetal or adult, appear to have similar patterns of gene expressions (clustered together at the bottom of the figure). Genes that are highly expressed in eye tissues fall in three clusters, shown with a black box. These clusters are displayed in more detail in Figure 4A, B and C.
Extended Data Fig. 5 Genes from the regions associated with RE (from Supplementary Table 3) that are particularly expressed in eye tissues, compared to non-ocular tissues.
These clusters are those highlighted in Supplementary Figure 3, but for the sake of clarity they are shown in transposed orientation compared to the previous figure (here genes in the y-axis and eye tissues in the x-axis), but same color codes as before. The dendrograms represent the degree of similarity observed for both tissues and gene expressions. The clusters are given in the order in which they were clustered together, from left to right: a) genes that are expressed more in other ocular tissues (fetal and adult) but much less in the adult retina. b) genes that are highly expressed in the retina and other ocular tissues, and c) genes that are expressed in the retina, but less in the other ocular tissues tested.
Extended Data Fig. 6 Results of the LD score regression analysis applied to specifically expressed genes on multiple tissue for the meta-analysis results.
Each point represents one tissue or cell line (along the x-axis) and the log10 value of the p-value for the enrichment of the meta-analysis results among genes expressed in these tissues. There were 205 tests carried out, one in each tissue and cell line, therefore only tissues with a correlation p-value< 0.00025 (Log_P > 3.6 in this figure), would have been significant after multiple testing. This condition was not fulfilled for any of the available tissues.
Extended Data Fig. 7 Mendelian randomization results on causality of IOP over refractive error. Single points in the graph represent coordinates determined by the effect of each specific SNP over IOP (x-axis, mmHg) and spherical equivalent (y-axis, Diopter units).
A total of 73 SNPs associated with IOP, but not directly associated with refractive error (that is p > 0.05) were selected as instruments. Values of associations with IOP were obtained from a meta-analysis of 139,555 European participants (Reference 50 in the manuscript) and the refractive error associations from 102,117 UK Biobank subjects. The lines represent the regression lines from each model, as specified in the figure legend. In some cases, these lines may not visible because they overlap (please refer to the values underneath the figure).
Extended Data Fig. 8 Venn’s Diagram of the number of SNPs considered in each of the stages of this study.
The different circles represent various stages, inclusion in the meta-analysis (blue), identification of significant loci (green), conditional analysis results identifying independent effects (red) and the total number of SNPs available for inclusion in prediction and heritability estimation in the independent (that is not part of the original meta-analysis) EPIC-Norfolk cohort (orange).
Extended Data Fig. 9 Prediction for the total number of SNPs and phenotypic variance explained as a function of GWAS sample size in future studies, based on the distribution of effects observed in the current meta-analysis.
The plot lines show the predicted relationship between the number of loci associated with refractive error (left vertical axis, blue line) and the variance they help explain (red line, right vertical axis), as a function of the sample size (x-axis) used in future GWAS or meta-analyses. These projections are consistent with the observed results, where an effective sample of 379,227 identified 904 independent signals after a conditional analysis, explaining 12–16% of refractive error variability.
Extended Data Fig. 10 The distribution of various natural selection test scores for SNPs associated with refractive error.
The values on the x-axis represent the ranking in terms of natural selection observed and the y-axis the density of that rank. The different tests shown are iHS, XP-EHH (CEU vs YRI), XP-EHH average score, XP-EHH maximum score and Tajima scores (black, green, red, blue and yellow respectively).
Supplementary information
Supplementary Information
Supplementary Note
Supplementary Tables
Supplementary Tables 1–26
Supplementary Data Set 1
Comparison of effect sizes illustrating the consistent effects observed in three of the major components of the meta-analysis.
Supplementary Data Set 2
LocusZoom Plots of association signals originating from autosomal regions and overlapping with genes indicated in the main manuscript. When available (not all SNPs are present and have linkage disequilibrium data available in the LocusZoom server files), the index SNP is highlighted, and the different colors denote LD with adjacent SNPs in that region. Colder colors usually denote independence (low LD). Occasionally, within the plot window, there will be different SNPs with stronger association compared to the index SNP, but their effects are sufficiently independent. The SNPs are annotated to the nearest protein coding gene, within a 250 kb interval.
Supplementary Data Set 3
Source Data for Fig. 1
Supplementary Data Set 4
Source Data for Fig. 2
Supplementary Data Set 5
Source Data for Extended Data Fig. 1
Supplementary Data Set 6
Source Data for Extended Data Fig. 2
Supplementary Data Set 7
Source Data for Extended Data Fig. 3
Supplementary Data Set 8
Source Data for Extended Data Fig. 4
Supplementary Data Set 9
Source Data for Extended Data Fig. 5
Supplementary Data Set 10
Source Data for Extended Data Fig. 6
Supplementary Data Set 11
Source Data for Extended Data Fig. 7
Supplementary Data Set 12
Information about Extended Data Fig. 8
Supplementary Data Set 13
Source Data for Extended Data Fig. 9
Supplementary Data Set 14
Source Data for Extended Data Fig. 10
Rights and permissions
About this article
Cite this article
Hysi, P.G., Choquet, H., Khawaja, A.P. et al. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat Genet 52, 401–407 (2020). https://doi.org/10.1038/s41588-020-0599-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-0599-0
This article is cited by
-
The influence of the environment and lifestyle on myopia
Journal of Physiological Anthropology (2024)
-
Diurnal retinal and choroidal gene expression patterns support a role for circadian biology in myopia pathogenesis
Scientific Reports (2024)
-
Ocular manifestations of renal ciliopathies
Pediatric Nephrology (2024)
-
Circadian rhythm, ipRGCs, and dopamine signalling in myopia
Graefe's Archive for Clinical and Experimental Ophthalmology (2024)
-
Signatures of adaptation in myopia-related genes on the sunlight exposure hypothesis
Journal of Physiological Anthropology (2023)