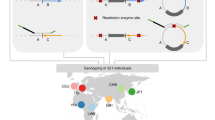
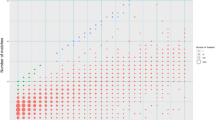
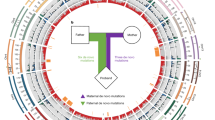
Abstract
Inversions play an important role in disease and evolution but are difficult to characterize because their breakpoints map to large repeats. We increased by sixfold the number (n = 1,069) of previously reported great ape inversions by using single-cell DNA template strand and long-read sequencing. We find that the X chromosome is most enriched (2.5-fold) for inversions, on the basis of its size and duplication content. There is an excess of differentially expressed primate genes near the breakpoints of large (>100 kilobases (kb)) inversions but not smaller events. We show that when great ape lineage-specific duplications emerge, they preferentially (approximately 75%) occur in an inverted orientation compared to that at their ancestral locus. We construct megabase-pair scale haplotypes for individual chromosomes and identify 23 genomic regions that have recurrently toggled between a direct and an inverted state over 15 million years. The direct orientation is most frequently the derived state for human polymorphisms that predispose to recurrent copy number variants associated with neurodevelopmental disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Strand-seq data aligned to GRCh38 and ape-specific composite files are available at zenodo, (https://doi.org/10.5281/zenodo.3818043); the PacBio and Bionano datasets are reported in Supplementary Tables 11 and 14; Supplementary data are available at GitHub (https://github.com/daewoooo/ApeInversion_paper); the PacBio and Bionano inversion callset are available at GitHub (https://github.com/daewoooo/ApeInversion_paper/tree/master/Supplementary_datasets).
Code availability
The primatR package is available at GitHub (https://github.com/daewoooo/primatR); the breakpointR package is available at GitHub (https://github.com/daewoooo/breakpointR) (devel branch); custom scripts are available at GitHub (https://github.com/daewoooo/ApeInversion_paper/tree/master/Custom_scripts); software releases at the publication date are available at Zenodo (https://doi.org/10.5281/zenodo.3556774).
References
Sturtevant, A. H. Genetic factors affecting the strength of linkage in Drosophila. Proc. Natl Acad. Sci. USA 3, 555–558 (1917).
Antonacci, F. et al. Characterization of six human disease-associated inversion polymorphisms. Hum. Mol. Genet. 18, 2555–2566 (2009).
Chaisson, M. J. P., Wilson, R. K. & Eichler, E. E. Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 16, 627–640 (2015).
Chaisson, M. J. P. et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 10, 1784 (2019).
Kidd, J. M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008).
Kidd, J. M. et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143, 837–847 (2010).
Sanders, A. D. et al. Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res. 26, 1575–1587 (2016).
Zody, M. C. et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 40, 1076–1083 (2008).
Vicente-Salvador, D. et al. Detailed analysis of inversions predicted between two human genomes: errors, real polymorphisms, and their origin and population distribution. Hum. Mol. Genet. 26, 567–581 (2017).
Giner-Delgado, C. et al. Evolutionary and functional impact of common polymorphic inversions in the human genome. Nat. Commun. 10, 4222 (2019).
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nat. Genet. 37, 727–732 (2005).
Kronenberg, Z. N. et al. High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 (2018).
Yunis, J. & Prakash, O. The origin of man: a chromosomal pictorial legacy. Science 215, 1525–1530 (1982).
Kehrer-Sawatzki, H., Sandig, C. A., Goidts, V. & Hameister, H. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet. Genome Res. 108, 91–97 (2005).
Kehrer-Sawatzki, H. et al. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes). Hum. Mutat. 25, 45–55 (2005).
Ventura, M. et al. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res. 22, 1036–1049 (2012).
Lucas Lledó, J. I. & Cáceres, M. On the power and the systematic biases of the detection of chromosomal inversions by paired-end genome sequencing. PLoS ONE 8, e61292 (2013).
Catacchio, C. R. et al. Inversion variants in human and primate genomes. Genome Res. 28, 910–920 (2018).
Alkan, C., Coe, B. P. & Eichler, E. E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12, 363–376 (2011).
Rasekh, M. E. et al. Discovery of large genomic inversions using long range information. BMC Genomics 18, 65 (2017).
Feuk, L. et al. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 1, e56 (2005).
Falconer, E. et al. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat. Methods 9, 1107–1112 (2012).
Sanders, A. D., Falconer, E., Hills, M., Spierings, D. C. J. & Lansdorp, P. M. Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat. Protoc. 12, 1151–1176 (2017).
Porubsky, D. et al. breakpointR: an R/Bioconductor package to localize strand state changes in Strand-seq data. Bioinformatics 36, 1260–1261 (2020).
Szamalek, J. M. et al. The chimpanzee-specific pericentric inversions that distinguish humans and chimpanzees have identical breakpoints in Pan troglodytes and Pan paniscus. Genomics 87, 39–45 (2006).
Sulovari, A. et al. Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc. Natl Acad. Sci. USA 116, 23243–23253 (2019).
Newman, T. L. et al. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 15, 1344–1356 (2005).
Shao, H. et al. npInv: accurate detection and genotyping of inversions using long read sub-alignment. BMC Bioinformatics 19, 261 (2018).
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012).
Cheng, Z. et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437, 88–93 (2005).
Sudmant, P. H. et al. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 23, 1373–1382 (2013).
Osborne, L. R. et al. A 1.5 million-base pair inversion polymorphism in families with Williams–Beuren syndrome. Nat. Genet. 29, 321–325 (2001).
Giglio, S. et al. Olfactory receptor–gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 68, 874–883 (2001).
Coe, B. P. et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 46, 1063–1071 (2014).
Vollger, M. R. et al. Long-read sequence and assembly of segmental duplications. Nat. Methods 16, 88–94 (2019).
Marques-Bonet, T. et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457, 877–881 (2009).
Ventura, M. et al. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res. 21, 1640–1649 (2011).
Spielmann, M., Lupiáñez, D. G. & Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 19, 453–467 (2018).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Brawand, D. et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011).
Sousa, A. M. M. et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017).
Pollen, A. A. et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756.e17 (2019).
Kanton, S. et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019).
Ghavi-Helm, Y. et al. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 51, 1272–1282 (2019).
Hey, J. Speciation and inversions: chimps and humans. Bioessays 25, 825–828 (2003).
Navarro, A. & Barton, N. H. Chromosomal speciation and molecular divergence—accelerated evolution in rearranged chromosomes. Science 300, 321–324 (2003).
Fishilevich, S. et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017, bax028 (2017).
Sohoni, S. et al. Elevated heme synthesis and uptake underpin intensified oxidative metabolism and tumorigenic functions in non-small cell lung cancer cells. Cancer Res. 79, 2511–2525 (2019).
Cáceres, M., Sullivan, R. T. & Thomas, J. W. A recurrent inversion on the eutherian X chromosome. Proc. Natl Acad. Sci. USA 104, 18571–18576 (2007).
Bailey, J. A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002).
Corbett-Detig, R. B. & Hartl, D. L. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8, e1003056 (2012).
Natri, H. M., Merilä, J. & Shikano, T. The evolution of sex determination associated with a chromosomal inversion. Nat. Commun. 10, 145 (2019).
Kong, A. et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467, 1099–1103 (2010).
Nuttle, X. et al. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature 536, 205–209 (2016).
Fuller, Z. L., Leonard, C. J., Young, R. E., Schaeffer, S. W. & Phadnis, N. Ancestral polymorphisms explain the role of chromosomal inversions in speciation. PLoS Genet. 14, e1007526 (2018).
Lozier, J. N. et al. The Chapel Hill hemophilia A dog colony exhibits a factor VIII gene inversion. Proc. Natl Acad. Sci. USA 99, 12991–12996 (2002).
Itsara, A. et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 84, 148–161 (2009).
Antonacci, F. et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 46, 1293–1302 (2014).
Mohajeri, K. et al. Interchromosomal core duplicons drive both evolutionary instability and disease susceptibility of the Chromosome 8p23.1 region. Genome Res. 26, 1453–1467 (2016).
Maggiolini, F. A. M. et al. Genomic inversions and GOLGA core duplicons underlie disease instability at the 15q25 locus. PLoS Genet. 15, e1008075 (2019).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Gel, B. et al. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32, 289–291 (2016).
Porubský, D. et al. Direct chromosome-length haplotyping by single-cell sequencing. Genome Res. 26, 1565–1574 (2016).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Sudmant, P. H. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010).
Lewis, P. O. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925 (2001).
Cleary, J. G. et al. Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J. Comput. Biol. 21, 405–419 (2014).
Porubsky, D. et al. Dense and accurate whole-chromosome haplotyping of individual genomes. Nat. Commun. 8, 1293 (2017).
Weirather, J. L. et al. Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 43, e116 (2015).
Tukiainen, T. et al. Landscape of X chromosome inactivation across human tissues. Nature 550, 244–248 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
We thank T. Brown for assistance in editing this manuscript. In addition, we thank S. Pääbo for generously providing the bonobo (Ulindi) and chimpanzee (Dorien) cell lines used in this study, along with H. Kaessmann and E. Leushkin for access to the ape RNA-seq data. We acknowledge the technical assistance provided by A. Pang and A. Hastie, who provided the Bionano inversion calls for NHPs. We also thank the European Molecular Biology Laboratory Genomics Core facility, particularly V. Benes and J. Zimmermann, for assistance with automating the Strand-seq library generation. This work was supported, in part, by grants from the National Institutes of Health (NIH; grant nos. HG002385 and HG010169 to E.E.E.). A.D.S. was supported by an Alexander von Humboldt Foundation Research Fellowship. P.H. was supported by the NIH Pathway to Independence Award (National Human Genome Research Institute, no. K99HG011041). A.S. was supported by the NIH Genome Training Grant (T32, no. HG000035-23). J.O.K. was supported by a European Research Council Consolidator grant (no. 773026). S.C. was supported by a National Health and Medical Research Council CJ Martin Biomedical Fellowship (no. 1073726). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
D.P., A.D.S. and E.E.E. designed the study, analyzed and interpreted the data, produced the figures and wrote the manuscript. A.D.S. and J.O.K. generated the Strand-seq libraries. W.H. analyzed the TADs and differential gene expression. P.H. and A.S. reconstructed the NHP phylogeny and helped with the statistical analysis. R.L., M.S., S.C., L.M., M.V. and F.A. provided validation of the inversion calls. S.C.M. and D.G. processed the PacBio data. T.M. and A.A.P. supported data analysis and interpretation.
Corresponding author
Ethics declarations
Competing interests
E.E.E. is on the scientific advisory board of DNAnexus.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–32, Note and Tables 4, 8 and 11–14
Supplementary Tables
Supplementary Tables 1–3, 5–7, 9 and 10
Rights and permissions
About this article
Cite this article
Porubsky, D., Sanders, A.D., Höps, W. et al. Recurrent inversion toggling and great ape genome evolution. Nat Genet 52, 849–858 (2020). https://doi.org/10.1038/s41588-020-0646-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-0646-x
This article is cited by
-
Inversion polymorphism in a complete human genome assembly
Genome Biology (2023)
-
Haplotype-resolved assemblies and variant benchmark of a Chinese Quartet
Genome Biology (2023)
-
Human-specific genetics: new tools to explore the molecular and cellular basis of human evolution
Nature Reviews Genetics (2023)
-
SVDSS: structural variation discovery in hard-to-call genomic regions using sample-specific strings from accurate long reads
Nature Methods (2023)
-
Inversions maintain differences between migratory phenotypes of a songbird
Nature Communications (2023)