Abstract
Parental epigenomes are established during gametogenesis. While they are largely reset after fertilization, broad domains of Polycomb repressive complex 2 (PRC2)-mediated formation of lysine 27–trimethylated histone H3 (H3K27me3) are inherited from oocytes in mice. How maternal H3K27me3 is established and inherited by embryos remains elusive. Here, we show that PRC1-mediated formation of lysine 119–monoubiquititinated histone H2A (H2AK119ub1) confers maternally heritable H3K27me3. Temporal profiling of H2AK119ub1 dynamics revealed that atypically broad H2AK119ub1 domains are established, along with H3K27me3, during oocyte growth. From the two-cell stage, H2AK119ub1 is progressively deposited at typical Polycomb targets and precedes H3K27me3. Reduction of H2AK119ub1 by depletion of Polycomb group ring finger 1 (PCGF1) and PCGF6—essential components of variant PRC1 (vPRC1)—leads to H3K27me3 loss at a subset of genes in oocytes. The gene-selective H3K27me3 deficiency is irreversibly inherited by embryos, causing loss of maternal H3K27me3-dependent imprinting, embryonic sublethality and placental enlargement at term. Collectively, our study unveils preceding dynamics of H2AK119ub1 over H3K27me3 at the maternal-to-zygotic transition, and identifies PCGF1/6–vPRC1 as an essential player in maternal epigenetic inheritance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All of the CUT&RUN and RNA-seq datasets generated in this study are summarized in Supplementary Table 6 and have been deposited to the Gene Expression Omnibus database under accession number GSE153496. The H2AK119ub1 ChIP-seq datasets of mESCs were from GSE119620 (ref. 41). The H3K27me3 and H3K4me3 ChIP-seq and whole-genome bisulfite sequencing datasets of FGOs were from GSE93941 (ref. 19). The H3K27me3 ChIP-seq datasets of 7-d GOs, MII oocytes, zygotes, early two-cell embryos, late two-cell embryos, inner cell masses of blastocysts and E6.5 epiblasts were from GSE76687 (ref. 18). The H3K27me3 CUT&RUN datasets of morula embryos were from GSE116713 (ref. 26). The H3K27me3 ChIP-seq datasets in mESCs were from GSE119620 (ref. 41). The RNA-seq datasets of Ring1a/1b KO FGOs and Dnmt3l matKO and Eed matKO morula embryos were from GSE132156 (ref. 56), GSE130115 (ref. 29) and GSE116713 (ref. 26), respectively. The PCGF1 ChIP-seq datasets in mESCs were from GSE119620 (ref. 41).
Code availability
The code developed for this study is available at https://github.com/Azusa-lab/Intergenerational-epigenetic-inheritance.
References
Eckersley-Maslin, M. A., Alda-Catalinas, C. & Reik, W.Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018).
Borsos, M. et al. Genome–lamina interactions are established de novo in the early mouse embryo. Nature 569, 729–733 (2019).
Burton, A. & Torres-Padilla, M. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief. Funct. Genomics 9, 444–454 (2010).
Burton, A. & Torres-Padilla, M. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 15, 723–735 (2014).
Xu, Q. & Xie, W. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol. 28, 237–253 (2018).
Probst, A. & Almouzni, G. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 27, 177–185 (2011).
Duffie, R. & Bourc’his, D. Parental epigenetic asymmetry in mammals. Curr. Top. Dev. Biol. 104, 293–328 (2013).
Feil, R. Epigenetic asymmetry in the zygote and mammalian development. Int. J. Dev. Biol. 53, 191–201 (2009).
Collombet, S. et al. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature 580, 142–146 (2020).
Flyamer, I. M. et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nat. Genet. 544, 110–114 (2017).
Du, Z. et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547, 232–235 (2017).
Ke, Y. et al. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170, 367–381.e20 (2017).
Liu, X. et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016).
Wang, C. et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 20, 620–631 (2018).
Xu, Q. et al. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat. Genet. 51, 844–856 (2019).
Zhang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016).
Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016).
Zheng, H. et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079 (2016).
Hanna, C. W. et al. MLL2 conveys transcription-independent H3K4 trimethylation in oocytes. Nat. Struct. Mol. Biol. 25, 73–82 (2018).
Sankar, A. et al. KDM4A regulates the maternal-to-zygotic transition by protecting broad H3K4me3 domains from H3K9me3 invasion in oocytes. Nat. Cell Biol. 22, 380–388 (2020).
Yeung, W. K. A. et al. Histone H3K9 methyltransferase G9a in oocytes is essential for preimplantation development but dispensable for CG methylation protection. Cell Rep. 27, 282–293.e4 (2019).
Tucci, V. et al. Genomic imprinting and physiological processes in mammals. Cell 176, 952–965 (2019).
Ferguson-Smith, A. C. Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12, 565–575 (2011).
Chen, Z. & Zhang, Y. Maternal H3K27me3-dependent autosomal and X chromosome imprinting. Nat. Rev. Genet. 21, 555–571 (2020).
Harris, C. et al. Conversion of random X-inactivation to imprinted X-inactivation by maternal PRC2. eLife 8, e44258 (2019).
Inoue, A., Chen, Z., Yin, Q. & Zhang, Y. Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev. 32, 1525–1536 (2018).
Inoue, A., Jiang, L., Lu, F., Suzuki, T. & Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 (2017).
Inoue, A., Jiang, L., Lu, F. & Zhang, Y. Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 31, 1927–1932 (2017).
Chen, Z., Inoue, A., Yin, Q., Zhang, C. & Zhang, Y. Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci. Adv. 5, eaay7246 (2019).
Hanna, C. W. et al. Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol. 20, 225 (2019).
Gaydos, L. J., Wang, W. & Strome, S. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345, 1515–1518 (2014).
Batista, R. A. & Köhler, C. Genomic imprinting in plants—revisiting existing models. Genes Dev. 34, 24–36 (2020).
Zenk, F. et al. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 357, 212–216 (2017).
Skvortsova, K., Iovino, N. & Bogdanovic, O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 19, 774–790 (2018).
Schuettengruber, B., Bourbon, H.-M., Di Croce, L. & Cavalli, G. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171, 34–57 (2017).
Aranda, S., Mas, G. & Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 1, e1500737 (2015).
Blackledge, N. P., Rose, N. R. & Klose, R. J. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol. 16, 643–649 (2015).
Barbour, H., Daou, S., Hendzel, M. & Affar, E. B. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat. Commun. 11, 5947 (2020).
Gao, Z. et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 (2012).
Blackledge, N. P. et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and Polycomb domain formation. Cell 157, 1445–1459 (2014).
Fursova, N. A. et al. Synergy between variant PRC1 complexes defines Polycomb-mediated gene repression. Mol. Cell 74, 1020–1036.e8 (2019).
Blackledge, N. P. et al. PRC1 catalytic activity is central to Polycomb system function. Mol. Cell 77, 857–874.e9 (2020).
Tamburri, S. et al. Histone H2AK119 mono-ubiquitination is essential for Polycomb-mediated transcriptional repression. Mol. Cell 77, 840–856.e5 (2020).
Tavares, L. et al. RYBP–PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678 (2012).
Almeida, M. et al. PCGF3/5–PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356, 1081–1084 (2017).
Cooper, S. et al. Targeting Polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 7, 1456–1470 (2014).
Cooper, S. et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 7, 13661 (2016).
Kalb, R. et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 21, 569–571 (2014).
Rose, N. R. et al. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. eLife 5, e18591 (2016).
Pintacuda, G. et al. hnRNPK recruits PCGF3/5–PRC1 to the Xist RNA B-repeat to establish Polycomb-mediated chromosomal silencing. Mol. Cell 68, 955–969.e10 (2017).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017).
Scelfo, A. et al. Functional landscape of PCGF proteins reveals both RING1A/B-dependent-and RING1A/B-independent-specific activities. Mol. Cell 74, 1037–1052.e7 (2019).
Van den Boom, V. et al. Non-canonical PRC1.1 targets active genes independent of H3K27me3 and Is essential for leukemogenesis. Cell Rep. 14, 332–346 (2016).
Zhou, Y., Romero-Campero, F. J., Gomez-Zambrano, A., Turck, F. & Calonje, M. H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 18, 69 (2017).
Loubiere, V., Papadopoulos, G. L., Szabo, Q., Martinez, A.-M. & Cavalli, G. Widespread activation of developmental gene expression characterized by PRC1-dependent chromatin looping. Sci. Adv. 6, eaax4001 (2020).
Du, Z. et al. Polycomb group proteins regulate chromatin architecture in mouse oocytes and early embryos. Mol. Cell 77, 825–839.e7 (2020).
Posfai, E. et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 26, 920–932 (2012).
Puschendorf, M. et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40, 411–420 (2008).
Tardat, M. et al. Cbx2 targets PRC1 to constitutive heterochromatin in mouse zygotes in a parent-of-origin-dependent manner. Mol. Cell 58, 157–171 (2015).
Eid, A. & Torres-Padilla, M. E. Characterization of non-canonical Polycomb repressive complex 1 subunits during early mouse embryogenesis. Epigenetics 11, 389–397 (2016).
De Vries, W. N. et al. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 26, 110–112 (2000).
Zepeda-Martinez, J. A. et al. Parallel PRC2/cPRC1 and vPRC1 pathways silence lineage-specific genes and maintain self-renewal in mouse embryonic stem cells. Sci. Adv. 6, eaax5692 (2020).
Endoh, M. et al. PCGF6–PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. eLife 6, e21064 (2017).
Stielow, B., Finkernagel, F., Stiewe, T., Nist, A. & Suske, G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 14, e1007193 (2018).
Farcas, A. M. et al. KDM2B links the Polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife 1, e00205 (2012).
Matoba, S. et al. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 23, 343–354.e5 (2018).
Dobrinić, P., Szczurek, A. T. & Klose, R. J. PRC1 drives Polycomb-mediated gene repression by controlling transcription initiation and burst frequency. Preprint at bioRxiv https://doi.org/10.1101/2020.10.09.333294 (2020).
Riising, E. M. et al. Gene silencing triggers Polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell 55, 347–360 (2014).
Yu, J.-R., Lee, C.-H., Oksuz, O., Stafford, J. M. & Reinberg, D. PRC2 is high maintenance. Genes Dev. 33, 903–935 (2019).
Wakayama, T. & Yanagimachi, R.Cloning of male mice from adult tail-tip cells. Nat. Genet. 22, 127–128 (1999).
Okae, H. et al. RNA sequencing-based identification of aberrant imprinting in cloned mice. Hum. Mol. Genet. 23, 992–1001 (2014).
Wang, L. Y. et al. Overcoming intrinsic H3K27me3 imprinting barriers improves post-implantation development after somatic cell nuclear transfer. Cell Stem Cell 27, 315–325.e5 (2020).
Inoue, K. et al. Loss of H3K27me3 imprinting in the Sfmbt2 miRNA cluster causes enlargement of cloned mouse placentas. Nat. Commun. 11, 2150 (2020).
Haig, D. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc. Biol. Sci. 264, 1657–1662 (1997).
Grossniklaus, U., Vielle-Calzada, J. P., Hoeppner, M. A. & Gagliano, W. B. Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280, 446–450 (1998).
Healy, E. et al. PRC2.1 and PRC2.2 synergize to coordinate H3K27 trimethylation. Mol. Cell 76, 437–452.e6 (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Robinson, J. et al. Integrative Genomics Viewer. Nat. Biotechnol. 29, 24–26 (2011).
Ramirez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Krueger, F. & Andrews, S. R. SNPsplit: allele-specific splitting of alignments between genomes with known SNP genotypes. F1000Res 5, 1479 (2016).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Yu, G., Wang, L.-G. & He, Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Acknowledgements
We thank all of the Koseki laboratory members for helpful discussion and the Kazusa DNA Research Institute for support with next-generation sequencing. The protein A-MNase for CUT&RUN was a kind gift from S. Henikoff. We appreciate T. Ishiuchi (Kyushu University), S. Ito (RIKEN) and S. Yamaguchi (Osaka University) for critical reading of the manuscript and K. Yamamoto and T. Yamamoto (RIKEN) for support as the young chief investigator program directors. This project was partly supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Leading Initiative for Excellent Young Researchers Grant (to A.I.), Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) (18H02359 to A.I. and 16H02622 to H.K.), Grants-in-Aid for Scientific Research on Innovative Areas (19H05754 to A.I. and 19H05745 to H.K.), Grant-in-Aid for Early-Career Scientists (19K17971 to C.K.), Grant-in-Aid for JSPS Fellows (20J21541 to R.H.), Japan Agency for Medical Research and Development (PRIME; JP18gm6110012 to A.I.), AMED-CREST (JP18gm0510016 to H.K.), the Uehara Memorial Foundation (to A.I.) and intramural grants within RIKEN, including the All-RIKEN ‘Epigenome Manipulation Project’ (to A.I. and H.K.).
Author information
Authors and Affiliations
Contributions
A.I. conceived the project and designed the experiments. H.M. analyzed the sequencing data. A.I., C.K., R.H. and M.K. performed the experiments. R.H. helped with the data analysis. A.I. and H.M. interpreted the data. H.K. provided the Pcgf1 and Pcgf6 floxed mouse lines. A.I. and H.M. wrote the manuscript with contributions from H.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Neil Brockdorff, Maxim Greenberg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of H2AK119ub1 distribution in mouse oocytes.
a, Scatter plots showing H2AK119ub1 enrichment (10 kb window) between our low-input CUT&RUN datasets using 250, 500, 1,000, 5,000 mouse embryonic stem cells (mESCs) and a public ChIP-seq dataset in mESCs41. Spearman correlation is also shown. b, Genome browser view of H2AK119ub1 in our low-input CUT&RUN datasets and a public ChIP-seq dataset of mESCs. c, Scatter plot showing the correlation between biological duplicate of H2AK119ub1 CUT&RUN in FGOs. d, Pie chart showing the percentage of H2AK119ub1 (H2Aub)-covered regions that overlap H3K27me3 (K27me3) or/and H3K4me3 (K4me3). e, Genome browser view showing local association of H2AK119ub1 and H3K4me3 at promoters in FGOs. The H3K4me3 and H3K27me3 ChIP-seq datasets are from19. f, Heatmap showing the enrichment of H2AK119ub1, H3K27me3, and H3K4me3 at promoters harboring H2AK119ub1 peaks in FGOs. Reads density (normalized RPKM with different scales) are plotted at ±5 kb of the transcription start sites (TSS). The promoters are clustered into 3 groups according to the relative enrichment of the histone PTMs. The H3K27me3 and H3K4me3 ChIP-seq datasets are from19. The rightmost column represents the expression levels of genes corresponding to individual rows in FGOs. The RNA-seq dataset is from16. g, Box plot showing gene expression levels of the 3 clusters of panel f. The middle lines in the boxes represent the medians. Box edges, the upper, and the lower whiskers indicate the interquartile range (IQR, from the 25th to 75th percentile), the largest value smaller than 1.5 x IQR above the 75th percentile, and the smallest value larger than 1.5 x IQR below the 25th percentile, respectively (n = 1, as biological replicates were combined). *** p-value = 1.43e-71 (two sided Mann-Whitney U test). h, MA plot of log2FC(KO/CTR) in gene expression of Ring1a/1b KO FGOs. Cluster C genes in panel f are highlighted by red dots. The percentages of up- and down-regulated differentially expressed genes (DEGs) and non-DEGs with the fold change>2 cutoff are indicated. i, Scatter plot showing the correlation between biological duplicates of H2AK119ub1 CUT&RUN in 7d-GOs. j, Additional genome browser views of H2AK119ub1 and H3K27me3 distributions in 7d-GOs and FGOs. The H3K27me3 ChIP-seq datasets are from18.
Extended Data Fig. 2 Characterization of allelic H2AK119ub1 distribution at the maternal-to-zygotic transition.
a, Scatter plot showing the correlations between biological duplicates of H2AK119ub1 CUT&RUN in individual samples. b, The ratios of SNP-containing reads assigned to the paternal and maternal genomes. Note that MII oocytes have almost no paternal SNP reads, supporting the validity of our SNP analysis pipeline. c, Averaged signal profiles of H2AK119ub1 at regions with different gene densities (gene number per 1 Mb sliding window with a 2kb step) in zygotes (n=1, as biological replicates were combined). The shaded areas represent the 95% confidence interval for the fitted LOESS curve. Note that the H2AK119ub1 enrichment at the paternal allele is negatively correlated with gene density. d, Representative images of anti-H2AK119ub1 and anti-H3.3 immunostaining analysis for fertilized oocytes. Dotted circles, the rims of fertilized oocytes. M, maternal genome. P, paternal genome. Scale bar, 20 µm. Ten fertilized oocytes were examined in each time point. e, Enlarged images of the paternal genomes in panel d. DAPI-dense loci are pointed by arrows. DAPI was shown in blue in the merged images. Scale bar, 10 µm. f, Genomic distribution of H2AK119ub1 peaks. M, maternal allele. P, paternal allele. Promoters represent the regions of ±2.5 kb around transcription start sites.
Extended Data Fig. 3 H2AK119ub1 dynamics and gene expression during the maternal-to-zygotic transition.
a, Genome browser views of H2AK119ub1 and H3K27me3 dynamics at gene deserts. MII, MII oocyte. 1C, 1-cell. E2C, early 2-cell. L2C, late 2-cell. Mor, morula. Bl, blastocyst. Epi, E6.5 epiblast. b, c, Heatmaps showing the H2AK119ub1 and H3K27me3 enrichment at promoters (b) and gene bodies (c) in the maternal allele of the indicated samples. Genes with few SNP reads [RPKM(H2AK119ub1) < 1 in all samples] were filtered out from this analysis. The gene expression levels are shown in the right RNA-seq heatmap. The H3K27me3 ChIP-seq datasets are from18. The RNA-seq datasets are from16. d, Pie chart showing the proportion of putative H3K27me3-dependent imprinted genes27 in Group A/B/C of the panel c. Genes with few SNP reads [RPKM(H2AK119ub1) < 1 in all samples] were filtered out from this analysis. The total number of genes in this pie chart is 62. e, Pie chart showing the proportion of typical Polycomb (PcG) target genes in Group A/B/C of the panel b. Genes with few SNP reads [RPKM(H2AK119ub1) < 1 in all samples] were filtered out from this analysis. The total number of genes in this pie chart is 1,405. f, Genome browser views of H2AK119ub1 and H3K27me3 dynamics at typical PcG targets. The genomic length of each view is indicated at the top. ESC, embryonic stem cells.
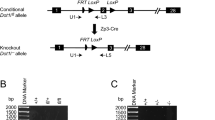
Extended Data Fig. 4 Generation of oocyte-specific Pcgf1/6 knockout mice.
a, The expression levels of the Pcgf family genes during oogenesis and preimplantation development. The RNA-seq datasets are from16. The exact RPKM values of Pcgf1 over 200 are indicated at the top of the bars. D10-and D14-GO, growing oocytes (GO) from postnatal day 10 (D10) and day 14 (D14) females. FGO, fully-grown oocytes. MII, MII-stage oocytes. ICM, inner cell mass of blastocysts. mESC, mouse embryonic stem cell. vPRC1, variant PRC1 components. cPRC1, canonical PRC1 components. b, Construct for targeted disruption of Pcgf1. Black boxes indicate the coding exons. Red arrows indicate genotyping primers. c, Genotyping of Pcgf1 and Pcgf6 flox alleles. The F2/R1 primer set was used to detect WT and flox alleles. d, Construct for targeted disruption of Pcgf6. e, Sanger sequencing to confirm the deletions of floxed exons in Pcgf1/6 KO FGOs. After cDNA preparation by reverse transcription of total RNA from WT or KO FGOs, the targeted regions were PCR amplified and sequenced. This confirmed that exons 2–7 (Pcgf1) and 2–3 (Pcgf6), which encode the Ring finger domains of PCGF1 and PCGF6, respectively, were successfully deleted in KO FGOs.
Extended Data Fig. 5 Characterization of Pcgf1/6 KO fully-grown oocytes (FGOs).
a, Heatmap showing the enrichment of H2AK119ub1 CUT&RUN signals in replicate 1 (not scaled) and 2 (scaled by spike-in chromatin). Reads densities are plotted at the peaks ±2 kb flanking regions. b, c, Scatter plots showing the correlations between biological duplicates of H2AK119ub1 (b) and H3K27me3 (c) CUT&RUN in CTR and KO FGOs. d, Heatmap showing the enrichment of H3K27me3 CUT&RUN signals. Reads density are plotted at the peaks ±2 kb flanking regions. e, Scatter plots showing the correlations between biological duplicates of RNA-seq in CTR and KO FGOs. f, Scatter plots showing the correlations between biological duplicates of H3K27me3 CUT&RUN in Pcgf1 and Pcgf6 single KO FGOs. g, Box plot showing the enrichment of PCGF1 binding at H3K27me3-lost, -intermediate, and -unchanged genes defined in Fig. 4e. PCGF1 ChIP-seq datasets in mESCs are from41. ***p = 1.8e-105 (two sided Mann-Whitney U test). h, Box plot showing CpG density at promoters of the 3 groups of genes. ***p = 3.9e-100 (two sided Mann-Whitney U test). i, Box plot showing H2AK119ub1 and H3K27me3 enrichment at gene bodies of the 3 groups in 7-day growing oocytes (7d-GOs). ***p = 2.3e-69 (two sided Mann-Whitney U test).
Extended Data Fig. 6 Preimplantation development of Pcgf1/6 maternal KO (matKO) embryos.
a, The averaged numbers of MII oocytes following superovulation. The numbers of females examined were 17 (CTR) and 14 (KO). Error bars, SD. b, Preimplantation development of Pcgf1/6 CTR and matKO embryos. The embryos that reached at the 2-cell, 4-cell, morula, and blastocyst stages in a timely fashion were counted at 24, 48, 72, and 96 hours post-fertilization (hpf), respectively. The numbers of 1-cell zygotes were set as 100%. The numbers of embryos examined are 127 (CTR) and 171 (matKO) from 6 biologically independent experiments. ***p < 0.001 (Chi-squired test). c, Representative images of preimplantation embryos at the indicated time points. The expected stages at these time points are indicated in parentheses. The arrowheads indicate embryos that had not reached the expected stages in a timely manner. Note that the other Pcgf1/6 matKO embryos form grossly normal blastocysts at 96 hpf. The experiment was repeated 6 times. Scale bar, 100 µm. d, Quantifications of H2AK119ub1 and H3K27me3 immunostaining analysis in Pcgf1/6 CTR and matKO embryos. The numbers of embryos examined (n) are indicated in Fig. 5a. The averaged signal intensity of CTR was set as 1.0 in each stage. Maternal pronuclei were quantified for 1-cell zygotes. Bars overlaid on the plots indicate mean. ***p < 0.001 (two-tailed Student’s t-test).
Extended Data Fig. 7 Characterization of Pcgf1/6 maternal KO (matKO) morula embryos.
a, b, Scatter plots showing the correlations between biological duplicates of RNA-seq in B6xPWK and PWKxB6 (a) and Pcgf1/6 CTR and matKO (b) morula embryos. c, The ratio of blastomeres showing the indicated numbers of Xist RNA clouds. Each bar represents an individual embryo. d, Box plot showing the relative expression of genes on individual maternal chromosomes between CTR and matKO morula embryos. Box edges, the upper, and the lower whiskers indicate the interquartile range (IQR, from the 25th to 75th percentile), the largest value smaller than 1.5 x IQR above the 75th percentile, and the smallest value larger than 1.5 x IQR below the 25th percentile, respectively (n=1, as biological replicates were combined). ***p < 2.2e-16 (One-way ANOVA test). e, Scatter plots showing the correlations between biological triplicates of H3K27me3 CUT&RUN in CTR and matKO morula embryos. f, Additional genome browser views of H3K27me3 distributions in Pcgf1/6 CTR and KO FGOs, and CTR and matKO morula embryos. Mat, maternal allele. Pat, paternal allele. No views in embryos are shown at Pnliprp2 and Gm32885 that have few SNPs. g, Heatmap showing the CUT&RUN signal enrichment of maternal H3K27me3 (scaled) in late 2-cell embryos. Reads density were plotted at the peaks ±2 kb flanking regions. h, Averaged signal profiles of maternal H3K27me3 (scaled) at their peaks ±2 kb flanking regions in late 2-cell embryos. i, Heatmap showing the enrichment of the H3K27me3 intensity in FGOs, late 2-cell, and morula embryos. The list and the order of genes are the same as Fig. 5d. Heatmaps for the corresponding paternal allele are also shown. The rightmost heatmap indicates H2AK119ub1 signal intensity at the maternal allele of wild-type preimplantation embryos.
Extended Data Fig. 8 Characterization of Pcgf1/6 maternal KO (matKO) embryos at E6.5.
a, Summary table of E6.5 dissection. b, Pictures of all of 4 CTR and 8 matKO litters at E6.5. Scale bar, 1 mm. c, Representative images of H3K27me3 immunostaining of E6.5 embryos. H3K27me3 spots are indicators of X chromosome inactivation (XCI). Note that single XCI and no XCI is observed in female and male embryos, respectively, in both CTR and matKO. This indicates that aberrant XCI in matKO embryos is restored by E6.5. Oct4-positive and -negative cells represent epiblast and extra-embryonic ectoderm, respectively. The number of embryos examined was 6 (CTR) and 9 (matKO) females and 9 (CTR) and 5 (matKO) males. Scale bar, 50 µm. d, Expression levels of cell lineage marker genes in CTR and matKO extraembryonic ectoderm (ExE) samples. RNA-seq datasets of wild-type epiblast (EPI), visceral endoderm (VE), and ExEs are from27. e, Correlation between biological replicates of RNA-seq samples.
Extended Data Fig. 9 Characterization of Pcgf1/6 double and respective single maternal KO (matKO) fetuses and placentae at term.
a, Summary table of Caesarean sections at E18.5. b, Experimental scheme of mixed embryo transfer. Genotyping of the ΔPcgf1 or ΔPcgf6 allele allows distinguishing between CTR and matKO fetuses. c, d, Placental weights (c) and body weights (d) of Pcgf1/6 CTR and matKO fetuses dissected from surrogated mothers at E19.5. Bars overlaid on the plots indicate mean±SD. The number of placentae and fetuses examined was 16 (CTR) and 8 (matKO) from 3 litters. ***p < 0.0001 (two-tailed Student’s t-test). e, Ratios of wild-type (WT), Pcgf1 heterozygous (Het), Pcgf6 Het, Pcgf1/6 double Het fetuses derived from WT females that had been mated with Pcgf1/6 double Het males. A total of 80 fetuses obtained from 9 litters were examined at E18.5. f, Placental weights of the indicated genotypes. Bars overlaid on the plots indicate mean±SD. p, two-tailed Student’s t-test. g, Summary table of Caesarean sections at E18.5 for Pcgf1 and Pcgf6 single matKO, respectively. h, i, j, The numbers of implantation (h), the litter sizes (i) and the placental weights (j) of the indicated matKO embryos. The numbers of litters and placentae examined are 11 and 85 (CTR), 10 and 24 (Pcgf1/6 matKO), 7 and 48 (Pcgf1 matKO), and 10 and 69 (Pcgf6 matKO), respectively. Bars overlaid on the plots indicate mean ± SD. ***p < 0.0001 (two-tailed Student’s t-test).
Supplementary information
Supplementary Tables
Supplementary Tables 1–7
Rights and permissions
About this article
Cite this article
Mei, H., Kozuka, C., Hayashi, R. et al. H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nat Genet 53, 539–550 (2021). https://doi.org/10.1038/s41588-021-00820-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00820-3
This article is cited by
-
Evaluating histone modification analysis of individual preimplantation embryos
BMC Genomics (2024)
-
The impact of selective HDAC inhibitors on the transcriptome of early mouse embryos
BMC Genomics (2024)
-
Conservation and divergence of canonical and non-canonical imprinting in murids
Genome Biology (2023)
-
Regulation, functions and transmission of bivalent chromatin during mammalian development
Nature Reviews Molecular Cell Biology (2023)
-
Casting histone variants during mammalian reproduction
Chromosoma (2023)