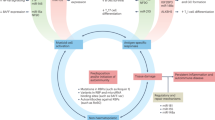
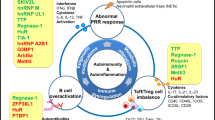
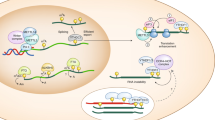
Abstract
RNA-binding proteins (RBPs) are essential for the development and function of the immune system. They interact dynamically with RNA to control its biogenesis and turnover by transcription-dependent and transcription-independent mechanisms. In this Review, we discuss the molecular mechanisms by which RBPs allow gene expression changes to occur at different speeds and to varying degrees, and which RBPs regulate the diversity of the transcriptome and proteome. These proteins are nodes for integration of transcriptional and signaling networks and are intimately linked to intermediary metabolism. They are essential components of regulatory feedback mechanisms that maintain immune tolerance and limit inflammation. The role of RBPs in malignancy and autoimmunity has led to their emergence as targets for the development of new therapeutic modalities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Marina Spence/Springer Nature.

Marina Spence/Springer Nature.

Marina Spence/Springer Nature.

Marina Spence/Springer Nature.
Similar content being viewed by others
References
Floor, S. N. & Doudna, J. A. Tunable protein synthesis by transcript isoforms in human cells. eLife 5, e10921 (2016).
Hombrink, P. et al. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17, 1467–1478 (2016).
Salerno, F., Paolini, N. A., Stark, R., von Lindern, M. & Wolkers, M. C. Distinct PKC-mediated posttranscriptional events set cytokine production kinetics in CD8+ T cells. Proc. Natl. Acad. Sci. USA 159, 201704227 (2017).
Turner, M., Galloway, A. & Vigorito, E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat. Immunol. 15, 484–491 (2014).
Hudson, W. H. & Ortlund, E. A. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 15, 749–760 (2014).
Chen, Y. G., Satpathy, A. T. & Chang, H. Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 18, 962–972 (2017).
Kafasla, P., Skliris, A. & Kontoyiannis, D. L. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat. Immunol. 15, 492–502 (2014).
Gerstberger, S., Hafner, M. & Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845 (2014). A compendium of RNA-binding proteins in the mammalian genome, indicating their structural and functional diversity.
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Brannan, K. W. et al. SONAR discovers RNA-binding proteins from analysis of large-scale protein-protein interactomes. Mol. Cell 64, 282–293 (2016).
He, C. et al. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell 64, 416–430 (2016).
Liepelt, A. et al. Identification of RNA-binding proteins in macrophages by interactome capture. Mol. Cell. Proteomics 15, 2699–2714 (2016).
Chang, X., Li, B. & Rao, A. RNA-binding protein hnRNPLL regulates mRNA splicing and stability during B-cell to plasma-cell differentiation. Proc. Natl. Acad. Sci. USA 112, E1888–E1897 (2015).
Yang, H., Duckett, C. S. & Lindsten, T. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol. Cell. Biol. 15, 6770–6776 (1995).
Techasintana, P. et al. The RNA-binding protein HuR posttranscriptionally regulates IL-2 homeostasis and CD4+ Th2 differentiation. ImmunoHorizons 1, 109–123 (2017).
Olejniczak, S. H. et al. Coordinated regulation of Cap-dependent translation and microRNA function by convergent signaling pathways. Mol. Cell. Biol. 36, 2360–2373 (2016).
Mallory, M. J. et al. Induced transcription and stability of CELF2 mRNA drives widespread alternative splicing during T-cell signaling. Proc. Natl. Acad. Sci. USA 112, E2139–E2148 (2015).
Tzachanis, D. et al. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol. 2, 1174–1182 (2001).
da Glória, V. G. et al. T cell activation regulates CD6 alternative splicing by transcription dynamics and SRSF1. J. Immunol. 193, 391–399 (2014).
Jeltsch, K. M. et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote TH17 differentiation. Nat. Immunol. 15, 1079–1089 (2014).
Diaz-Muñoz, M. D. et al. The RNA-binding protein HuR is essential for the B cell antibody response. Nat. Immunol. 16, 415–425 (2015).
Van Nostrand, E.L. et al. A large-scale binding and functional map of human RNA binding proteins. Preprint available at https://www.biorxiv.org/content/early/2017/08/23/179648 (2017).
Cieśla, J. Metabolic enzymes that bind RNA: yet another level of cellular regulatory network? Acta Biochim. Pol. 53, 11–32 (2006).
White, M. R. & Garcin, E. D. The sweet side of RNA regulation: glyceraldehyde-3-phosphate dehydrogenase as a noncanonical RNA-binding protein. Wiley Interdiscip. Rev. RNA 7, 53–70 (2016).
Jia, J. et al. Protection of extraribosomal RPL13a by GAPDH and dysregulation by S-nitrosylation. Mol. Cell 47, 656–663 (2012).
Chang, C.-H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016).
Salerno, F., Guislain, A., Cansever, D. & Wolkers, M. C. TLR-mediated innate production of IFN-γ by CD8+ T cells is independent of glycolysis. J. Immunol. 196, 3695–3705 (2016).
Hodge, D. L. et al. IFN-γ AU-rich element removal promotes chronic IFN-γ expression and autoimmunity in mice. J. Autoimmun. 53, 33–45 (2014).
Sakai, S. et al. CD4 T-cell-derived IFN-γ plays a minimal role in control of pulmonary Mycobacterium tuberculosis infection and must be actively repressed by PD-1 to prevent lethal disease. PLoS Pathog. 12, e1005667 (2016).
Panas, M. D., Ivanov, P. & Anderson, P. Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215, 313–323 (2016).
Scheu, S. et al. Activation of the integrated stress response during T helper cell differentiation. Nat. Immunol. 7, 644–651 (2006).
Díaz-Muñoz, M. D. et al. Tia1 dependent regulation of mRNA subcellular location and translation controls p53 expression in B cells. Nat. Commun. 8, 530 (2017).
Antonicka, H. & Shoubridge, E. A. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep 10, 920–932 (2015).
Hubstenberger, A. et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157 (2017). This study used flow cytometry to isolate processing bodies and analyze their makeup by proteomics.
Simsek, D. et al. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169, 1051–1065 (2017).
Horvilleur, E. et al. A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia 28, 1092–1102 (2014).
Shi, Z. et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67, 71–83 (2017).
Zinder, J. C. & Lima, C. D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 31, 88–100 (2017).
Pefanis, E. & Basu, U. RNA exosome regulates AID DNA mutator activity in the B cell genome. Adv. Immunol 127, 257–308 (2015).
Laffleur, B., Basu, U. & Lim, J. RNA exosome and non-coding RNA-coupled mechanisms in AID-mediated genomic alterations. J. Mol. Biol. 429, 3230–3241 (2017).
Kawaguchi, Y. et al. SRSF1-3 contributes to diversification of the immunoglobulin variable region gene by promoting accumulation of AID in the nucleus. Biochem. Biophys. Res. Commun. 485, 261–266 (2017).
Collart, M. A. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA 7, 438–454 (2016).
Treiber, T. et al. A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol. Cell 66, 270–284 (2017).
Batra, R. et al. RNA-binding protein CPEB1 remodels host and viral RNA landscapes. Nat. Struct. Mol. Biol. 23, 1101–1110 (2016).
Ivshina, M., Alexandrov, I. M., Vertii, A., Doxsey, S. & Richter, J. D. CPEB regulation of TAK1 synthesis mediates cytokine production and the inflammatory immune response. Mol. Cell. Biol. 35, 610–618 (2015).
Shin, J., Paek, K. Y., Ivshina, M., Stackpole, E. E. & Richter, J. D. Essential role for non-canonical poly(A) polymerase GLD4 in cytoplasmic polyadenylation and carbohydrate metabolism. Nucleic Acids Res 45, 6793–6804 (2017).
Mayr, C. Regulation by 3′-untranslated regions. Annu. Rev. Genet. 51, 171–194 (2017).
Beisang, D., Reilly, C. & Bohjanen, P. R. Alternative polyadenylation regulates CELF1/CUGBP1 target transcripts following T cell activation. Gene 550, 93–100 (2014).
Gruber, A. R. et al. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat. Commun. 5, 5465 (2014).
Berkovits, B. D. & Mayr, C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367 (2015).
Lim, J. et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159, 1365–1376 (2014).
Gutiérrez-Vázquez, C. et al. 3′ uridylation controls mature microRNA turnover during CD4 T-cell activation. RNA 23, 882–891 (2017).
Kozlowski, E. et al. The RNA uridyltransferase Zcchc6 is expressed in macrophages and impacts innate immune responses. PLoS One 12, e0179797 (2017).
Morgan, M. et al. mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 548, 347–351 (2017).
Elbarbary, R. A. et al. Tudor-SN-mediated endonucleolytic decay of human cell microRNAs promotes G1/S phase transition. Science 356, 859–862 (2017).
Roignant, J.-Y. & Soller, M. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 33, 380–390 (2017).
Vu, L. P. et al. The N 6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med 23, 1369–1376 (2017).
Li, H.-B. et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 (2017). References 58 and 59 offer clear demonstrations of roles of RNA methylation in immune cell development and function.
Schaub, A. & Glasmacher, E. Splicing in immune cells—mechanistic insights and emerging topics. Int. Immunol. 29, 173–181 (2017).
Kelemen, O. et al. Function of alternative splicing. Gene 514, 1–30 (2013).
Shukla, S. et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 (2011).
Wong, J. J.-L. et al. Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment. Nat. Commun. 8, 15134 (2017).
Marina, R. J. et al. TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. EMBO J. 35, 335–355 (2016).
Sharma, S. et al. Acetylation-dependent control of global poly(A) RNA degradation by CBP/p300 and HDAC1/2. Mol. Cell 63, 927–938 (2016).
Movassat, M. et al. Coupling between alternative polyadenylation and alternative splicing is limited to terminal introns. RNA Biol. 13, 646–655 (2016).
Enders, A. et al. Zinc-finger protein ZFP318 is essential for expression of IgD, the alternatively spliced Igh product made by mature B lymphocytes. Proc. Natl. Acad. Sci. USA 111, 4513–4518 (2014).
Pioli, P. D., Debnath, I., Weis, J. J. & Weis, J. H. Zfp318 regulates IgD expression by abrogating transcription termination within the Ighm/Ighd locus. J. Immunol. 193, 2546–2553 (2014).
Haneklaus, M., O’Neil, J. D., Clark, A. R., Masters, S. L. & O’Neill, L. A. J. The RNA-binding protein Tristetraprolin (TTP) is a critical negative regulator of the NLRP3 inflammasome. J. Biol. Chem. 292, 6869–6881 (2017).
Kurosaki, T. & Maquat, L. E. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 129, 461–467 (2016).
Middleton, R. et al. IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol. 18, 51 (2017).
Ni, T. et al. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res. 44, 6817–6829 (2016).
Cho, V. et al. The RNA-binding protein hnRNPLL induces a T cell alternative splicing program delineated by differential intron retention in polyadenylated RNA. Genome Biol. 15, R26 (2014).
Monzon-Casanova, E. et al. The RNA-binding protein PTBP1 is necessary for B cell selection in germinal center. Nat. Immunol. (in the press).
Wong, J. J.-L. et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154, 583–595 (2013).
Rentas, S. et al. Musashi-2 attenuates AHR signalling to expand human haematopoietic stem cells. Nature 532, 508–511 (2016).
Stumpo, D. J. et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 114, 2401–2410 (2009).
Fang, J. et al. Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat. Immunol. 18, 236–245 (2017).
Kristiansen, T. A. et al. Cellular barcoding links B-1a B cell potential to a fetal hematopoietic stem cell state at the single-cell level. Immunity 45, 346–357 (2016).
Zhou, Y. et al. Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J. Exp. Med. 212, 569–580 (2015).
Tijchon, E. et al. Tumor suppressors BTG1 and BTG2 regulate early mouse B-cell development. Haematologica 101, e272–e276 (2016).
Dolezal, E. et al. The BTG2-PRMT1 module limits pre-B cell expansion by regulating the CDK4-Cyclin-D3 complex. Nat. Immunol. 18, 911–920 (2017).
Inoue, T. et al. CNOT3 contributes to early B cell development by controlling Igh rearrangement and p53 mRNA stability. J. Exp. Med. 212, 1465–1479 (2015).
Yang, C.-Y. et al. Interaction of CCR4-NOT with EBF1 regulates gene-specific transcription and mRNA stability in B lymphopoiesis. Genes Dev. 30, 2310–2324 (2016).
Galloway, A. et al. RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science 352, 453–459 (2016).
Vogel, K. U., Bell, L. S., Galloway, A., Ahlfors, H. & Turner, M. The RNA-binding proteins Zfp36l1 and Zfp36l2 enforce the thymic β-selection checkpoint by limiting DNA damage response signaling and cell cycle progression. J. Immunol. 197, 2673–2685 (2016). References 85 and 86 uncover a conserved regulation of quiescence through suppression of G1-S-phase progression factors by the ZFP36l1 and ZFP36l2 RNA-binding proteins.
Galloway, A. & Turner, M. Cell cycle RNA regulons coordinating early lymphocyte development. Wiley Interdiscip. Rev. RNA 8, e1419 (2017).
DeMicco, A. et al. Lymphocyte lineage-specific and developmental stage specific mechanisms suppress cyclin D3 expression in response to DNA double strand breaks. Cell Cycle 15, 2882–2894 (2016).
Fu, M. & Blackshear, P. J. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 17, 130–143 (2017).
Maeda, K. & Akira, S. Regulation of mRNA stability by CCCH-type zinc-finger proteins in immune cells. Int. Immunol. 29, 149–155 (2017).
Mino, T. et al. Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161, 1058–1073 (2015).
Wawro, M., Kochan, J., Krzanik, S., Jura, J. & Kasza, A. Intact NYN/PIN-like domain is crucial for the degradation of inflammation-related transcripts by ZC3H12D. J. Cell. Biochem. 118, 487–498 (2017).
Kapoor, N. et al. Transcription factors STAT6 and KLF4 implement macrophage polarization via the dual catalytic powers of MCPIP. J. Immunol. 194, 6011–6023 (2015).
Garg, A. V. et al. MCPIP1 endoribonuclease activity negatively regulates interleukin-17-mediated signaling and inflammation. Immunity 43, 475–487 (2015).
Monin, L. et al. MCPIP1/Regnase-1 restricts IL-17A- and IL-17C-dependent skin inflammation. J. Immunol. 198, 767–775 (2017).
Ebner, F. et al. The RNA-binding protein tristetraprolin schedules apoptosis of pathogen-engaged neutrophils during bacterial infection. J. Clin. Invest. 127, 2051–2065 (2017).
Andrianne, M. et al. Tristetraprolin expression by keratinocytes controls local and systemic inflammation. JCI Insight 2, 92979 (2017).
Tarling, E. J. et al. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J. Clin. Invest. 127, 3741–3754 (2017).
Newman, R. et al. Maintenance of the marginal-zone B cell compartment specifically requires the RNA-binding protein ZFP36L1. Nat. Immunol. 18, 683–693 (2017).
Chen, J. et al. Posttranscriptional gene regulation of IL-17 by the RNA-binding protein HuR is required for initiation of experimental autoimmune encephalomyelitis. J. Immunol. 191, 5441–5450 (2013).
Gubin, M. M. et al. Conditional knockout of the RNA-binding protein HuR in CD4+ T cells reveals a gene dosage effect on cytokine production. Mol. Med. 20, 93–108 (2014).
Techasintana, P., Davis, J. W., Gubin, M. M., Magee, J. D. & Atasoy, U. Transcriptomic-wide discovery of direct and indirect HuR RNA targets in activated CD4+ T cells. PLoS One 10, e0129321 (2015).
DeMicco, A. et al. B cell-intrinsic expression of the HuR RNA-binding protein is required for the T cell-dependent immune response in vivo. J. Immunol. 195, 3449–3462 (2015).
La Porta, J., Matus-Nicodemos, R., Valentín-Acevedo, A. & Covey, L. R. The RNA-binding protein, polypyrimidine tract-binding protein 1 (PTBP1) is a key regulator of CD4 T cell activation. PLoS One 11, e0158708 (2016).
Jin, Z., Liang, F., Yang, J. & Mei, W. hnRNP I regulates neonatal immune adaptation and prevents colitis and colorectal cancer. PLoS Genet. 13, e1006672 (2017).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015).
Hawse, W. F., Boggess, W. C. & Morel, P. A. TCR signal strength regulates Akt substrate specificity to induce alternate murine Th and T regulatory cell differentiation programs. J. Immunol. 199, 589–597 (2017).
Meininger, I. et al. Alternative splicing of MALT1 controls signalling and activation of CD4+ T cells. Nat. Commun. 7, 11292 (2016).
Martinez, N. M. et al. Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation. Genes Dev. 29, 2054–2066 (2015).
Mallory, M. J. et al. Signal- and development-dependent alternative splicing of LEF1 in T cells is controlled by CELF2. Mol. Cell. Biol. 31, 2184–2195 (2011).
Ajith, S. et al. Position-dependent activity of CELF2 in the regulation of splicing and implications for signal-responsive regulation in T cells. RNA Biol. 13, 569–581 (2016).
Sedlyarov, V. et al. Tristetraprolin binding site atlas in the macrophage transcriptome reveals a switch for inflammation resolution. Mol. Syst. Biol. 12, 868 (2016).
Clark, A. R. & Dean, J. L. E. The control of inflammation via the phosphorylation and dephosphorylation of tristetraprolin: a tale of two phosphatases. Biochem. Soc. Trans. 44, 1321–1337 (2016).
Tiedje, C. et al. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. 44, 7418–7440 (2016).
Smallie, T. et al. Dual-specificity phosphatase 1 and tristetraprolin cooperate to regulate macrophage responses to lipopolysaccharide. J. Immunol. 195, 277–288 (2015).
Ross, E. A. et al. Dominant suppression of inflammation via targeted mutation of the mRNA destabilizing protein tristetraprolin. J. Immunol. 195, 265–276 (2015).
Patial, S. et al. Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. Proc. Natl. Acad. Sci. USA 113, 1865–1870 (2016).
Newton, R., Shah, S., Altonsy, M. O. & Gerber, A. N. Glucocorticoid and cytokine crosstalk: feedback, feedforward, and co-regulatory interactions determine repression or resistance. J. Biol. Chem. 292, 7163–7172 (2017).
Shah, S., Mostafa, M. M., McWhae, A., Traves, S. L. & Newton, R. Negative feed-forward control of tumor necrosis factor (TNF) by tristetraprolin (ZFP36) is limited by the mitogen-activated protein kinase phosphatase, dual-specificity phosphatase 1 (DUSP1): implications for regulation by glucocorticoids. J. Biol. Chem. 291, 110–125 (2016).
Tang, T. et al. Macrophage responses to lipopolysaccharide are modulated by a feedback loop involving prostaglandin E2, dual specificity phosphatase 1 and tristetraprolin. Sci. Rep. 7, 4350 (2017).
Wang, K.-T. et al. Functional regulation of Zfp36l1 and Zfp36l2 in response to lipopolysaccharide in mouse RAW264.7 macrophages. J. Inflamm. (Lond.) 12, 42 (2015).
Zhang, Q. et al. New insights into the RNA-binding and E3 ubiquitin ligase activities of Roquins. Sci. Rep. 5, 15660 (2015).
Maruyama, T. et al. Roquin-2 promotes ubiquitin-mediated degradation of ASK1 to regulate stress responses. Sci. Signal. 7, ra8 (2014).
Cano, F., Rapiteanu, R., Sebastiaan Winkler, G. & Lehner, P. J. A non-proteolytic role for ubiquitin in deadenylation of MHC-I mRNA by the RNA-binding E3-ligase MEX-3C. Nat. Commun. 6, 8670 (2015).
Tenekeci, U. et al. K63-ubiquitylation and TRAF6 pathways regulate mammalian P-body formation and mRNA decapping. Mol. Cell 62, 943–957 (2016).
Protter, D. S. W. & Parker, R. Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016).
Hu, G. et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat. Cell Biol. 17, 930–942 (2015). This paper links RNA decay to the regulation of autophagy and mTOR-dependent processes.
Manfrini, N. et al. High levels of eukaryotic initiation factor 6 (eIF6) are required for immune system homeostasis and for steering the glycolytic flux of TCR-stimulated CD4+ T cells in both mice and humans. Dev. Comp. Immunol. 77, 69–76 (2017).
Chesney, J. et al. An inducible gene product for 6-phosphofructo-2-kinase with an AU-rich instability element: role in tumor cell glycolysis and the Warburg effect. Proc. Natl. Acad. Sci. USA 96, 3047–3052 (1999).
Zhang, J. et al. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell 19, 66–80 (2016).
Bayeva, M. et al. mTOR regulates cellular iron homeostasis through tristetraprolin. Cell Metab. 16, 645–657 (2012).
Yoshinaga, M. et al. Regnase-1 maintains iron homeostasis via the degradation of transferrin receptor 1 and prolyl-hydroxylase-domain-containing protein 3 mRNAs. Cell Rep. 19, 1614–1630 (2017).
Vasudevan, S. & Peltz, S. W. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 7, 1191–1200 (2001).
Dejure, F. R. et al. The MYC mRNA 3′-UTR couples RNA polymerase II function to glutamine and ribonucleotide levels. EMBO J. 36, 1854–1868 (2017).
Huang, W. et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 528, 517–522 (2015). This paper provides an excellent example of noncoding RNA interactions with RBP determining cell fate and function.
Xing, Z., Wang, S. & Tran, E. J. Characterization of the mammalian DEAD-box protein DDX5 reveals functional conservation with S. cerevisiae ortholog Dbp2 in transcriptional control and glucose metabolism. RNA 23, 1125–1138 (2017).
Castello, A. et al. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 (2016).
Niphakis, M. J. et al. A global map of lipid-binding proteins and their ligandability in cells. Cell 161, 1668–1680 (2015).
Clingman, C. C. et al. Allosteric inhibition of a stem cell RNA-binding protein by an intermediary metabolite. eLife 3, 693 (2014).
Ramiscal, R. R. et al. Attenuation of AMPK signaling by ROQUIN promotes T follicular helper cell formation. eLife 4, e08698 (2015).
Dvinge, H., Kim, E., Abdel-Wahab, O. & Bradley, R. K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430 (2016).
Tummala, H. et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest. 125, 2151–2160 (2015).
Dhanraj, S. et al. Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN). J. Med. Genet. 52, 738–748 (2015).
Stuart, B. D. et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat. Genet. 47, 512–517 (2015).
Hodson, D. J. et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat. Immunol. 11, 717–724 (2010).
Liu, Y. et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 49, 1211–1218 (2017).
Kesarwani, M. et al. Targeting c-FOS and DUSP1 abrogates intrinsic resistance to tyrosine-kinase inhibitor therapy in BCR-ABL-induced leukemia. Nat. Med. 23, 472–482 (2017).
Vantourout, P. et al. Immunological visibility: posttranscriptional regulation of human NKG2D ligands by the EGF receptor pathway. Sci. Transl. Med. 6, 231ra49 (2014).
Schmiedel, D. et al. The RNA binding protein IMP3 facilitates tumor immune escape by downregulating the stress-induced ligands ULPB2 and MICB. eLife 5, 727 (2016).
Galarza-Muñoz, G. et al. Human epistatic interaction controls IL7R splicing and increases multiple sclerosis risk. Cell 169, 72–84 (2017).
Acknowledgements
We thank E. Glasmacher for allowing us to discuss her unpublished findings, and our many colleagues who have provided feedback and support during our preparation of this manuscript. We also thank E. Werbenko for providing images for Fig. 2. The Biotechnology and Biological Sciences Research Council, the Medical Research Council, Bloodwise and Wellcome support work in M.T.'s laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Turner, M., Díaz-Muñoz, M.D. RNA-binding proteins control gene expression and cell fate in the immune system. Nat Immunol 19, 120–129 (2018). https://doi.org/10.1038/s41590-017-0028-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-017-0028-4
This article is cited by
-
Regulation of inflammatory diseases via the control of mRNA decay
Inflammation and Regeneration (2024)
-
RNA-binding proteins potentially regulate the alternative splicing of cell cycle-associated genes in proliferative diabetic retinopathy
Scientific Reports (2024)
-
RNA processing mechanisms contribute to genome organization and stability in B cells
Oncogene (2024)
-
Post-transcriptional checkpoints in autoimmunity
Nature Reviews Rheumatology (2023)
-
RBP–RNA interactions in the control of autoimmunity and autoinflammation
Cell Research (2023)