Abstract
Dengue virus is a major pathogen, and severe infections can lead to life-threatening dengue hemorrhagic fever. Dengue virus exists as four serotypes, and dengue hemorrhagic fever is often associated with secondary heterologous infections. Antibody-dependent enhancement (ADE) may drive higher viral loads in these secondary infections and is purported to result from antibodies that recognize dengue virus but fail to fully neutralize it. Here we characterize two antibodies, 2C8 and 3H5, that bind to the envelope protein. Antibody 3H5 is highly unusual as it not only is potently neutralizing but also promotes little if any ADE, whereas antibody 2C8 has strong capacity to promote ADE. We show that 3H5 shows resilient binding in endosomal pH conditions and neutralizes at low occupancy. Immunocomplexes of 3H5 and dengue virus do not efficiently interact with Fcγ receptors, which we propose is due to the binding mode of 3H5 and constitutes the primary mechanism of how ADE is avoided.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. Structure factors and final refined coordinates of crystal structures have been deposited in the RCSB PDB (https://www.rcsb.org/pdb/) under the accession codes 6FLA (3H5 complex form 1), 6FLB (3H5 form2), and 6FLC (2C8).
References
Bhatt, S. et al. The global distribution and burden of dengue. Nature 496, 504–507 (2013).
Halstead, S. B. Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus Res. 60, 421–467 (2003).
Halstead, S. B. & O’Rourke, E. J. Antibody-enhanced dengue virus infection in primate leukocytes. Nature 265, 739–741 (1977).
Guzman, M. G., Alvarez, M. & Halstead, S. B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 158, 1445–1459 (2013).
Pierson, T. C. et al. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1, 135–145 (2007).
Oliphant, T. et al. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 80, 12149–12159 (2006).
Dejnirattisai, W. et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat. Immunol. 17, 1102–1108 (2016).
Priyamvada, L. et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl Acad. Sci. USA 113, 7852–7857 (2016).
Hadinegoro, S. R. et al. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N. Engl. J. Med. 373, 1195–1206 (2015).
Screaton, G., Mongkolsapaya, J., Yacoub, S. & Roberts, C. New insights into the immunopathology and control of dengue virus infection. Nat. Rev. Immunol. 15, 745–759 (2015).
Mukhopadhyay, S., Kuhn, R. J. & Rossmann, M. G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3, 13–22 (2005).
Modis, Y., Ogata, S., Clements, D. & Harrison, S. C. Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 (2004).
Zhang, X. et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20, 105–110 (2013).
Zhao, H. et al. Structural basis of Zika virus-specific antibody protection. Cell 166, 1016–1027 (2016).
Austin, S. K. et al. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS Pathog. 8, e1002930 (2012).
Dowd, K. A., Jost, C. A., Durbin, A. P., Whitehead, S. S. & Pierson, T. C. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 7, e1002111 (2011).
Zhang, X. et al. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl Acad. Sci. USA 110, 6795–6799 (2013).
Fibriansah, G. et al. Structural changes in dengue virus when exposed to a temperature of 37°C. J. Virol. 87, 7585–7592 (2013).
Beltramello, M. et al. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8, 271–283 (2010).
Lai, C.-Y. et al. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J. Virol. 82, 6631–6643 (2008).
de Alwis, R. et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl Acad. Sci. USA 109, 7439–7444 (2012).
Fibriansah, G. et al. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat. Commun. 6, 6341 (2015).
Fibriansah, G. et al. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol. Med. 6, 358–371 (2014).
Dejnirattisai, W. et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177 (2015).
Rouvinski, A. et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520, 109–113 (2015).
Dejnirattisai, W. et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328, 745–748 (2010).
Lok, S.-M. The interplay of dengue virus morphological diversity and human antibodies. Trends Microbiol. 24, 284–293 (2016).
Robinson, L. N. et al. Structure-guided design of an anti-dengue antibody directed to a non-immunodominant epitope. Cell 162, 493–504 (2015).
Gentry, M. K., Henchal, E. A., McCown, J. M., Brandt, W. E. & Dalrymple, J. M. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am. J. Trop. Med. Hyg. 31, 548–555 (1982).
Morens, D. M., Halstead, S. B. & Marchette, N. J. Profiles of antibody-dependent enhancement of dengue virus type 2 infection. Microb. Pathog. 3, 231–237 (1987).
Stiasny, K., Allison, S. L., Schalich, J. & Heinz, F. X. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76, 3784–3790 (2002).
Stanfield, R. L., Zemla, A., Wilson, I. A. & Rupp, B. Antibody elbow angles are influenced by their light chain class. J. Mol. Biol. 357, 1566–1574 (2006).
Gromowski, G. D. & Barrett, A. D. T. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 366, 349–360 (2007).
Edeling, M. A. et al. Potent dengue virus neutralization by a therapeutic antibody with low monovalent affinity requires bivalent engagement. PLoS. Pathog. 10, e1004072 (2014).
Nybakken, G. E. et al. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437, 764–769 (2005).
Robbiani, D. F. et al. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell 169, 597–609.e11 (2017).
Gallichotte, E. N. et al. Human dengue virus serotype 2 neutralizing antibodies target two distinct quaternary epitopes. PLoS. Pathog. 14, e1006934 (2018).
Fibriansah, G. et al. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349, 88–91 (2015).
Lok, S.-M. et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15, 312–317 (2008).
Dowd, K. A., Mukherjee, S., Kuhn, R. J. & Pierson, T. C. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J. Virol. 88, 11726–11737 (2014).
Midgley, C. M. et al. Structural analysis of a dengue cross-reactive antibody complexed with envelope domain III reveals the molecular basis of cross-reactivity. J. Immunol 188, 4971–4979 (2012).
Li, J. et al. Structural and functional characterization of a cross-reactive dengue virus neutralizing antibody that recognizes a cryptic epitope. Structure 26, 51–59.e4 (2018).
Wang, J. et al. A human bi-specific antibody against Zika virus with high therapeutic potential. Cell 171, 229–241.e15 (2017).
Oliphant, T. et al. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11, 522–530 (2005).
Heinz, F. X. & Stiasny, K. The antigenic structure of Zika virus and its relation to other flaviviruses: implications for infection and immunoprophylaxis. Microbiol. Mol. Biol. Rev. 81, e00055-16 (2017).
Sukupolvi-Petty, S. et al. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J. Virol. 84, 9227–9239 (2010).
Shrestha, B. et al. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS. Pathog. 6, e1000823 (2010).
Siegrist, C. A. et al. Determinants of infant responses to vaccines in presence of maternal antibodies. Vaccine 16, 1409–1414 (1998).
Siber, G. R. et al. Interference of immune globulin with measles and rubella immunization. J. Pediatr. 122, 204–211 (1993).
Sundström, C. & Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 17, 565–577 (1976).
Lozzio, C. B. & Lozzio, B. B. Cytotoxicity of a factor isolated from human spleen. J. Natl Cancer Inst. 50, 535–538 (1973).
Nettleship, J. E. et al. A pipeline for the production of antibody fragments for structural studies using transient expression in HEK 293T cells. Protein Expr. Purif. 62, 83–89 (2008).
Midgley, C. M. et al. An in-depth analysis of original antigenic sin in dengue virus infection. J. Virol. 85, 410–421 (2011).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Winter, G., Lobley, C. M. C. & Prince, S. M. Decision making in xia2. Acta Crystallogr. D 69, 1260–1273 (2013).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012).
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007).
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Acknowledgements
This work was supported by the Wellcome Trust, UK; the Newton-Medical Research Council, UK (MR/NO12658/2); the National Institute for Health Research Oxford Biomedical Research Centre funding scheme; the Thailand National Center for Genetic Engineering; and Biotechnology, the Academy of Finland; the Office of the Higher Education Commission and Mahidol University under the National Research Universities Initiative; the Research Chair Grant from the National Science and Technology Development Agency (NSTDA), Thailand; and Faculty of Medicine Siriraj Hospital, Mahidol University, Grant Number (IO)R015936005. The OPIC electron microscopy facility was founded by a Wellcome Trust JIF award (060208/Z/00/Z) and is supported by a Wellcome Trust equipment grant (093305/Z/10/Z). The Wellcome Trust is also acknowledged for providing administrative support (Grant 075491/Z/04). G.R.S. is a Wellcome Trust senior investigator (095541/Z/11/Z and 203224/Z/16/Z). M.R. and A.F. were supported by Wellcome Trust fellowships (204703/Z/16/Z and 080721/Z/06/Z). We thank A. Siebert for support with electron microscopy; T. Prommool and C. Tawilert for scale-up and purification of monoclonal antibodies; P. Keelapang for good advice on NT and ADE assays; and D. Bitto for assistance with cryo-EM sample preparation. We also thank the staff of the Diamond Light Source (proposal MX8423), and ESRF, K. Harlos, and T. Walter, for assistance with crystallization; and D. Stuart for scientific discussion.
Author information
Authors and Affiliations
Contributions
Experiments were conceived and designed by A.F., M.R., W.D., J.M., J.T.H., J.M.G., and G.R.S. Experiments were performed by A.F., M.R., W.D., P.S., W.W., K.C., T.D., A.C., and C.M.M. The data were analyzed by A.F., M.R., W.D., C.M.M., J.M., J.T.H., J.M.G., and G.R.S.. The paper was written by A.F., M.R., J.M., J.T.H., J.M.G. and G.R.S. C.P., W.K. and P.M. provided antibodies.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 ADE in K562 cells and Fcγ receptor subtype dependence.
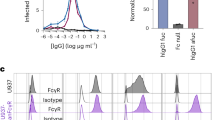
a,b, Enhancement properties of 3H5 (orange) and 2C8 (blue) in K562 cells. DENV2 strains NGC and 16681 (as indicated) were incubated with antibody before infecting K562 cells for enhancement assays. Supernatants of K562 cultures were harvested after 4 d and titrated onto Vero cells for a focus-forming assay. c,d, Effect of Fcγ receptor blocking on enhancement. Anti-FcγR antibodies (as indicated) were used to block Fc receptors during incubation of antibody–virus immunecomplexes with U937 cells or K562 cells to assess routes of enhancement. Supernatants of the cultures were harvested after 4 d and titrated onto Vero cells for a focus-forming assay.
Supplementary Figure 2 Comparison of the enhancement and neutralization properties of humanized 2C8-LALA.
a–d, LALA mutations were introduced into humanized 2C8 (Hu2C8) and enhancement (a,b) and neutralization (c,d) characteristics were assessed for DENV2/NGC and DENV2/16681 as indicated. In all experiments, DENV2 strains NGC and 16681 were incubated with antibody before infecting Vero or U937 cells for neutralization or enhancement assays, respectively. Supernatants of U937 cultures were harvested after 4 d and titrated onto Vero cells for a focus-forming assay. The data are shown as means ± s.e.m. from three independent experiments.
Supplementary Figure 3 Representative electron density.
a, Stereo diagram of the interaction between the EDIII FG loop (blue) and the 3H5 heavy chain (gray) and light chain (green). The interfacial waters involved in water-mediated hydrogen bonds are shown in red. The hydrogen bonds and salt bridges are marked with black dashed lines. b, Stereo diagram of the interaction between the N-terminal region of EDIII (blue) and the 2C8 Fab heavy chain (gray) and light chain (teal).
Supplementary Figure 4 CDRs of 2C8 and 3H5.
a, 2C8 variable domains (VL in light green and VH in dark green) bound to EDIII (blue) in cartoon representation. The complementarity-determining regions (CDRs) for the light chain (L1–L3) and heavy chain (H1–H3) are indicated. b, 3H5 variable domains (VL in light orange and VH in orange) bound to EDIII (blue). The CDRs are indicated. c, CDR-L1 of 3H5 intimately interacts with the surface of EDIII (blue) with the key residue F32 (shown as sticks) located at a central position.
Supplementary Figure 5 Comparison of EDIII residues targeted by 3H5 and quaternary epitope antibodies 2D22 and C8-EDE.
The general architecture of EDIII is shown on the left and important regions are labeled. Residues engaged by the lateral ridge antibody 3H5 are displayed as orange sticks. Residues on EDIII that are included in the epitope of quaternary antibodies 2D22 and C8 EDE are shown as green and yellow sticks, respectively.
Supplementary Figure 6 Cryo-electron microscopy analysis of DENV-2 incubated with 2C8 Fabs at room temperature.
a, Representative cryo-EM micrograph of DENV-2 particles incubated with 2C8 Fabs at room temperature showing virions that are partially occupied with Fabs. b, Comparison of the radial density profiles of particles incubated with 2C8 at room temperature (black curve) and at 37 °C (red curve) showing a distinct increase in Fab density (indicated by the white arrow). Radial density profiles were calculated from 2D averages of centered particles using IMAGIC.
Supplementary information
Supplementary Text, Figures and Tables
Supplementary Figures 1–6 and Supplementary Tables 1–3
Rights and permissions
About this article
Cite this article
Renner, M., Flanagan, A., Dejnirattisai, W. et al. Characterization of a potent and highly unusual minimally enhancing antibody directed against dengue virus. Nat Immunol 19, 1248–1256 (2018). https://doi.org/10.1038/s41590-018-0227-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0227-7
This article is cited by
-
Discovery of B-cell epitopes for development of dengue vaccines and antibody therapeutics
Medical Microbiology and Immunology (2022)
-
Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins
Nature Communications (2021)
-
Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor
Nature Medicine (2020)
-
Flat-lying antibody prevents disease enhancement
Nature Immunology (2018)