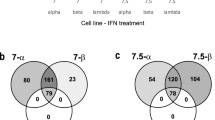
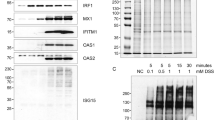
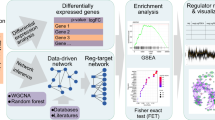
Abstract
Interferon-stimulated genes (ISGs) form the backbone of the innate immune system and are important for limiting intra- and intercellular viral replication and spread. We conducted a mass-spectrometry-based survey to understand the fundamental organization of the innate immune system and to explore the molecular functions of individual ISGs. We identified interactions between 104 ISGs and 1,401 cellular binding partners engaging in 2,734 high-confidence interactions. 90% of these interactions are unreported so far, and our survey therefore illuminates a far wider activity spectrum of ISGs than is currently known. Integration of the resulting ISG-interaction network with published datasets and functional studies allowed us to identify regulators of immunity and processes related to the immune system. Given the extraordinary robustness of the innate immune system, this ISG network may serve as a blueprint for therapeutic targeting of cellular systems to efficiently fight viral infections.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The MS-based proteomics data were deposited at the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository under following accession codes: PXD012274, PXD012275, PXD012276, PXD012277 and PXD012278. The protein interactions from this publication have been submitted to the IMEx consortium (http://www.imexconsortium.org) through IntAct and has been assigned the identifier IM-26691.
References
Schoggins, J. W. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Hayakawa, S. et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat. Immunol. 12, 37–44 (2011).
Li, M. M. et al. TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP). PLoS Pathog. 13, e1006145 (2017).0
Supek, F., Bosnjak, M., Skunca, N. & Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011).
Tokarev, A. et al. Stimulation of NF-κB activity by the HIV restriction factor BST2. J. Virol. 87, 2046–2057 (2013).
Galao, R. P., Le Tortorec, A., Pickering, S., Kueck, T. & Neil, S. J. Innate sensing of HIV-1 assembly by tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12, 633–644 (2012).
Wang, I. H., Burckhardt, C. J., Yakimovich, A., Morf, M. K. & Greber, U. F. The nuclear export factor CRM1 controls juxta-nuclear microtubule-dependent virus transport. J. Cell Sci. 130, 2185–2195 (2017).
Tripathi, S. et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18, 723–735 (2015).
de Wilde, A. H. et al. A kinome-wide small interfering RNA screen identifies proviral and antiviral host factors in severe acute respiratory syndrome coronavirus replication, including double-stranded RNA-activated protein kinase and early secretory pathway proteins. J. Virol. 89, 8318–8333 (2015).
York, A., Hutchinson, E. C. & Fodor, E. Interactome analysis of the influenza A virus transcription/replication machinery identifies protein phosphatase 6 as a cellular factor required for efficient virus replication. J. Virol. 88, 13284–13299 (2014).
Ma, J. et al. Structural and functional study of apoptosis-linked gene-2·heme-binding protein 2 interactions in HIV-1 production. J. Biol. Chem. 291, 26670–26685 (2016).
Long, J. S. et al. Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature 529, 101–104 (2016).
Zhao, K. et al. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J. Immunol. 196, 1209–1217 (2016).
Pichlmair, A. et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487, 486–490 (2012).
Rusinova, I. et al. Interferomev2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41, D1040–D1046 (2013).
Goffinet, C. Cellular antiviral factors that target particle infectivity of HIV-1. Curr. HIV Res. 14, 211–216 (2016).
Fogeron, M. L. et al. LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nat. Commun. 4, 1531 (2013).
White, M. J., Roife, D. & Gomer, R. H. Galectin-3 binding protein secreted by breast cancer cells inhibits monocyte-derived fibrocyte differentiation. J. Immunol. 195, 1858–1867 (2015).
Xiong, Y. et al. Annexin II receptor induces apoptosis independent of Annexin II. Apoptosis 18, 925–939 (2013).
Ito, K., Takahashi, A., Morita, M., Suzuki, T. & Yamamoto, T. The role of the CNOT1 subunit of the CCR4–NOT complex in mRNA deadenylation and cell viability. Protein Cell 2, 755–763 (2011).
Rodriguez-Gil, A. et al. The CCR4–NOT complex contributes to repression of major histocompatibility complex class II transcription. Sci. Rep. 7, 3547 (2017).
Chapat, C. et al. hCAF1/CNOT7 regulates interferon signalling by targeting STAT1. EMBO J. 32, 688–700 (2013).
Bar, I. et al. Knockout mice reveal a role for P2Y6 receptor in macrophages, endothelial cells, and vascular smooth muscle cells. Mol. Pharmacol. 74, 777–784 (2008).
Hacker, H., Tseng, P. H. & Karin, M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 11, 457–468 (2011).
Le May, N. et al. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116, 541–550 (2004).
Holze, C. et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat. Immunol. 19, 130–140 (2018).
Pichlmair, A. et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12, 624–630 (2011).
Reuther, P. et al. Generation of a variety of stable Influenza A reporter viruses by genetic engineering of the NS gene segment. Sci. Rep. 5, 11346 (2015).
Chase, G. P. et al. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS Pathog. 7, e1002187 (2011).
Riss, T. L. & Moravec, R. A. Use of multiple assay endpoints to investigate the effects of incubation time, dose of toxin, and plating density in cell-based cytotoxicity assays. Assay Drug Dev. Technol. 2, 51–62 (2004).
Gebhardt, A. et al. mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3. Nat. Commun. 6, 8192 (2015).
Hubner, N. C. et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 (2010).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Hein, M. Y. et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015).
Keilhauer, E. C., Hein, M. Y. & Mann, M. Accurate protein complex retrieval by affinity enrichment mass spectrometry (AE–MS) rather than affinity purification mass spectrometry (AP-MS). Mol. Cell Proteomics 14, 120–135 (2015).
Hothorn, T., Bretz, F. & Westfall, P. Simultaneous inference in general parametric models. Biom. J. 50, 346–363 (2008).
Benjamini, Y. & Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001).
Cox, J. & Mann, M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12 (2012).
Acknowledgements
We acknowledge the innate immunity laboratory for critical discussions and suggestions. We thank K. Mayr, I. Paron and G. Sowa for maintaining mass spectrometers and the MPI-B core facility for support. We further thank G. Kochs for the FluAV mini replicon and A549 cells, G. Zimmer for VSV-Luc and A. Merits for SFV-gLuc and SFV-NanoLuc. We also thank M. Feulner for technical assistance. The work in the authors’ laboratories was supported by the Max-Planck Free Floater program, ERC starting grant (ERC-StG iVIP, 311339), ERC consolidator grant (ERC-CoG ProDAP, 817798), Infect-Era and the German Federal Ministry of Education and Research (ERASE), the German Research Foundation (PI 1084/2, PI 1084/3, P1083/4 TRR179 and TRR237) to A.P. and (MS 632/15) to M.S. This work was also supported by NIH grant R01-AI091707 (to C.M.R.).
Author information
Authors and Affiliations
Contributions
P.H., C.U., V.B. B.K., W.M.S., P.S., L.B., H.H.H and A.M. conducted experiments. P.H. and A.S. analyzed data. M.S. and J.W.S. contributed critical reagents. P.H., M.S., M.M., C.M.R., A.S, P.S. and A.P. designed the experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Integrated supplementary information
Supplementary Figure 1 Bait selection and testing bait functionality.
(a) ISG proteins ranked by average antiviral activity in published gain of function virus replications studies. ISGs selected for AE-LC-MS/MS experiments are highlighted (blue circles). Top 10 ISGs showing highest and lowest antiviral activity are listed. (b) Validation of antiviral activity of selected ISGs in HEK-FlpIn TREX cells. ISG expression was induced by increasing concentrations of doxycycline (Dox) for 24h followed by infection with VSV-expressing firefly luciferase (VSV-Luc; MOI=0.01). Graphs show luciferase activity 18h post infection. Lysates were used to determine protein expression levels of the ISG transgene (HA), VSV-G and β-Tubulin by immunoblot analysis. The histogram shows average luciferase activity ±s.d.. n = 3 independent samples, one representative experiment of three is shown. * p-value ≤ 0,0002, ** p-value ≤ 0,0001, ns: not significant. One-way ANOVA with Dunnett’s multiple comparisons test. One representative experiment of three is shown.
Supplementary Figure 2 AE-LC-MS/MS procedure and identified interactions.
(a) AE-LC-MS/MS workflow and data analysis strategy. (b) The number of already reported and novel interactions identified by AE-LC-MS/MS experiments in this study. (c) The number of known and novel interactions for selected ISG baits, colors as in (b). (d) 293T cells were transfected for plasmids expressing SII-HA-tagged control (GFP, THYN1) or bait proteins (OASL, LGP2, RTCB, DDX60, CD74) together with Myc-tagged prey proteins (LARP7, IFI16, C14ORF166, eIF3e). Cell lysates of HEK293T cells were used for SII-immunoprecipitation and probed with antibodies against HA or Myc. One representative blot of three is shown. (e, f) Binary matrix of interactions between ISG baits (columns) and proteins (rows) with reported functions in NF-kB signaling (e) and in energy metabolism (f).
Supplementary Figure 3 Identification of ISG–ISG interactions and FluAV vRNP interactors.
(a) Binary matrix of ISG-ISG interactions identified in this study. Only ISGs listed in Supplementary Table 1 were considered. Bait ISG (columns), Prey ISG (rows). (b) BST2-centric NF-kB subnetwork. ISG baits (large circles) interacting with BST2 (red triangle) plus the other bait interactors (small balls) implicated in NF-kB signaling (based on GO BP annotations). (c) Identified ISG interactors with known functions in viral processes. Size and color intensity correspond to the number of interacting ISG baits (shown in callout boxes). (d, e) Co-immunoprecipitation of SII-HA-tagged control (THYN1, IFIT3) or bait proteins (NS4A4A, P2RY6, TNFSF13B, UNC93B1, VAMP5, CD74, and MAP3K14) and endogenous proteins (CSE1L, KPNB1). Cell lysates of 293T cells were used for SII-immunoprecipitation and probed with antibodies against HA-tag, CSE1L or KPNB1. One representative blot of three is shown. (f) Identification of FluAV vRNP-binding proteins by AE-LC-MS/MS analysis. A549 cells were infected with SII-tagged viral polymerase subunit 2 (PB2) expressing FluAV or untagged virus and anti-SII precipitates were analyzed by AE-LC-MS/MS. Volcano plot shows the average enrichment of a protein in PB2 pulldown compared to the control enrichments and the corresponding p-value (N = 4, two tailed t-test, unadjusted) with significantly enriched cellular (red) and viral (blue) proteins (permutation-based FDR = 0.05, S0 = 1, n = 4)
Supplementary Figure 4 Identification of LGALS3BP as an immunomodulatory protein.
(a) Co-immunoprecipitation of N-terminal-renilla-tagged LGALS3BP with non-transfected control (Mock), SII-HA-tagged control (GFP), or bait proteins (BCL2L14, FBXO6, MXA, SAMHD1, TNFRSF10A, VAMP5) in 293T cells. The graph shows mean ±s.d. relative luminescence unites (RLU) of streptavidin co-immunoprecipitated renilla-LGALS3BP normalized to the corresponding input renilla expression. Graph is based on technical triplicates (dots) of one representative biological repeat of three. * p-value < 0.01, ** p-value ≤ 0.001. Immunoblot analysis shows the enrichment of SII-HA-tagged proteins following SII-immunoprecipitation, probed with a HA-specific antibody. (b-e) HeLa cells were treated with siRNA against LGALS3BP, ATP6V0D1, ADAM17 or control siRNA (siCtrl) for 24h. The cells were (b) infected with FluAV-Ren (MOI=0.05), (c) infected with VSV expressing firefly luciferase (VSV-Luc), (d) transfected with plasmids that encode for the FluAV replication complex driving luciferase expression, (e) left untreated. (b, c, e) 24 h, (c) 36 h later (b) renilla, (c, d) firefly luciferase or (e) cell titer glow activity were measured. Histograms show average ±s.d. of three (b-d) or nine (e) technical replicates, representative experiments of at least three independent biological experiments are shown. ** p-value < 0.001, *** p-value < 0.0001, ns: not significant based on one way ANOVA with Dunnett’s multiple comparisons test. (f, g) Proteome analysis of HeLa cells treated with siRNA targeting LGALS3BP or Control (siScr) for 24h and subsequently infected with FluAV (MOI=0.05). (f) Protein expression in siLGALS3BP-treated FluAV-infected HeLa cells in comparison to siScr. Significantly enriched proteins are highlighted in red (two-tailed Welch’s t-test, FDR ≤ 0.01, S0 = 0.1, n = 4 independent measurements). (g) Top 5 GOBP terms enriched among the proteins significantly changed in (f) (Fisher’s exact test, Benjamini-Hochberg FDR ≤ 0.05, enrichment factor ≥ 2). (h) Proteome changes in LGALS3BP- (X axis) and ATP6V0D1- (Y axis) depleted HeLa cells in comparison to the non-targeting siRNA control (siScr). Red dots: ISGs upregulated in LGALS3BP-depleted cells. (i) Top 10 GOBP terms enriched among the proteins changed in (h). Histogram displays enrichment factors representing the mean LFQ based protein abundance distribution for each significant category, relative to the overall LFQ abundance distribution in the proteome. (Benjamini-Hochberg FDR ≤ 0.05).
Supplementary Figure 5 Functional effects of protein complex targeting.
(a) Selected protein complexes (based on GOCC) targeted by individual ISGs (Fisher’s exact test, Benjamini-Hochberg FDR ≤ 0.001, enrichment factor > 3). Shown protein complexes have ≤ 1 interactions of protein complex members to other ISGs in the network. Bar shows –log10 p-value of enrichment of the protein complex by the ISG against the background. (b) SII co-immunoprecipitation of endogenous CNOT1 with transfected SII-HA-tagged IFIT3 (control) or ANXA2R in 293T cells. One representative blot of three is shown. (c) HeLa-FlpIn TREX cells expressing the indicated transgene were left untreated or treated with doxycycline (1µg/ml) and at the same time infected with SFV-gLuc (MOI: 0.2). The graph shows mean luciferase activity ±s.d. of four independent samples 36h after infection. One representative experiment of three independent biological repeats is shown. p-values: Two way ANOVA with Tukey’s multiple comparisons test. * p < 0.0001, ns: not significant. (d) HeLa FlpIn TREX ANXA2R or Ctrl (IFIT3) cells were treated with doxycycline or not and cell viability was tested by resazurin reduction assay after the indicated time point. 12 individual samples per condition and timepoint were measured. One representative experiment of three independent biological repeats is shown. p-values: Two way ANOVA with Tukey’s multiple comparisons test. * p < 0.0001 between Mock and +Dox condition, ns: not significant. (e, f) As (c) but VSV-Luc (MOI: 1) was used for infection. (e) Mean luciferase activity ±s.d. of quadruplicate measurements 6h after infection. (f) Cell viability as measured by resazurine conversion assay 6h after infection. p-values: as (c), four independent samples of one representative experiment of three independent biological repeats is shown. (g) Confocal microscopy images of ANXAR2 and IFIT3 in HeLa FlpIn TREX cells. Doxycycline-treated ANXAR2 or control (IFIT3)-expressing HeLa-FlpIn TREX cells were fixed 24h after induction and immunostained for CNOT1 and HA-tagged proteins. Scale bars, 10 μm. n = 2 independent experiments. (h-j) Proteomic analysis of cells depleted for CNOT1 or overexpressing ANXA2R. (h) HeLa-FlpIn TREX cells were treated with siRNAs against CNOT1 or control (siScr) and the total proteome was analyzed by LC-MS/MS 48h later. Volcano plot shows all identified proteins. In blue: significantly up or downregulated proteins (student’s t-test, FDR ≤ 0.05, S0 = 0.05, n = four independent measurements). Proteins that are also significantly regulated in the same direction upon ANXA2R induction are highlighted in red, yellow or green. (i, j) HeLa-FlpIn TREX ANXA2R or control (IFIT3) cells were left unstimulated or stimulated with doxycycline and total proteome was analysed by LC-MS/MS 12h, 18h and 24h later. (i) Volcano plot showing proteome changes in doxycycline treated vs untreated HeLa-FlpIn TREX ANXA2R cells 24 hours post induction. Significantly changing (both in comparison to the no Dox control and to IFIT3 induction) proteins are highlighted in red (general linear model hypothesis test, Benjamini-Yekutieli adjusted p-value ≤ 0.001), and proteins significantly changing in (h) are further highlighted in blue. (j) Density plots showing changes of protein abundance distribution upon ANXA2R or control (IFIT3) induction. Proteins were grouped according to their change in CNOT1-depleted conditions shown in (h): unchanged upon CNOT1 depletion: grey, downregulated: blue, upregulated: pink. Numbers on top of each panel show p-values of two-sided K-S test for similarities between proteins co-regulated in CNOT1-depleted and ANXA2R- or control- (IFIT3) expressing conditions. ns: not significant.
Supplementary Figure 6 Activity of P2RY6.
(a, b) HEK-FlpIn TREX P2RY6 cells were stimulated with the indicated concentration of doxycycline (Dox) and infected with the indicated viruses. Bar plots show mean luciferase expression ±s.d. of doxycycline treated cells infected with VSV (MOI = 0.01) for 20h (a) and FluAV (MOI = 0.05) for 24h (b). Values were normalised to the non-doxycycline treated condition. Lysates in (a) were used for immunoblotting against HA-P2RY6, VSV-G and β-tubulin. Graphs show mean ±s.d. of three technical replicates, one representative experiment of three independent biological repeats is shown. (c) HEK-FlpIn TREX GFP cells were treated or not with doxycycline (Dox) for 20h and an additional 8h with the P2RY6 ligand MRS2957, as indicated. Cells were infected with SFV-NanoLuc (MOI: 0.01) for 24h and luciferase activity was measured. Histogram shows mean relative luciferase activity of quadruplicate measurements ±s.d. normalized to the uninduced control. (d, e) Proteome analysis of HEK-FlipIn TREX cells after doxycycline (Dox; 1µg/ml) induced P2RY6 expression for 24h. (d) Volcano plot shows average protein expression in doxycycline induced compared to uninduced HEK-FlipIn TREX cells (x-axis) and the transformed p-value for each protein (y-axis; two tailed t-test). Red dots: significant enriched proteins, curved dotted line separates significant enriched proteins from background population (black dots) (two-tailed t-test permutation based FDR < 0.01, S0 = 0.5, n = 6 independent measurements). p-values: One-way ANOVA with Dunnett’s multiple comparisons test. One representative experiment of three is shown. (e) Functional enrichment annotation analysis of significantly changed proteins compared to protein background population (Fisher’s exact test, Benjamini-Hochberg FDR < 0.05, enrichment factor > 2). Pathways derived from Panther database (http://www.pantherdb.org): AP1 pathway = Inflammation mediated by chemokine and cytokine signaling pathway → activator protein-1; JUN pathway = Gonadotropin-releasing hormone receptor pathway → JUN). (f) HEK 293 cells were co-transfected with a NF-KB-firefly luciferase reporter construct, an EIF1a-renilla control and increasing amounts of P2RY6 for 20h. GFP plasmid was used to adjust amounts of DNA for transfection. Cells were either treated or not with the P2RY6 specific ligand (MRS 2957, 250 nM) for additional 8h. Graph shows relative mean luciferase/renilla values ±s.d. of three replicates in relation to GFP control transfection. One representative experiment of three independent biological repeats is shown. (g) Control HEK-FlpIn TREX cells and HEK-FlpIn TREX MAP3K14 cells were infected with RVFV (Clone 13) for 18h, MOI 0.1 or not infected and used for AE-LC-MS/MS experiments. Analysis was performed as described in Fig. 6d and material and methods.
Supplementary information
Rights and permissions
About this article
Cite this article
Hubel, P., Urban, C., Bergant, V. et al. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat Immunol 20, 493–502 (2019). https://doi.org/10.1038/s41590-019-0323-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0323-3
This article is cited by
-
RNA binding protein SAMD4: current knowledge and future perspectives
Cell & Bioscience (2023)
-
Telomere-to-mitochondria signalling by ZBP1 mediates replicative crisis
Nature (2023)
-
UBE2O reduces the effectiveness of interferon-α via degradation of IFIT3 in hepatocellular carcinoma
Cell Death & Disease (2023)
-
The ubiquitination landscape of the influenza A virus polymerase
Nature Communications (2023)
-
mRNA 3’UTR lengthening by alternative polyadenylation attenuates inflammatory responses and correlates with virulence of Influenza A virus
Nature Communications (2023)