Abstract
Ectopic expression of defined transcription factors can force direct cell-fate conversion from one lineage to another in the absence of cell division. Several transcription factor cocktails have enabled successful reprogramming of various somatic cell types into induced neurons (iNs) of distinct neurotransmitter phenotype. However, the nature of the intermediate states that drive the reprogramming trajectory toward distinct iN types is largely unknown. Here we show that successful direct reprogramming of adult human brain pericytes into functional iNs by Ascl1 and Sox2 encompasses transient activation of a neural stem cell-like gene expression program that precedes bifurcation into distinct neuronal lineages. During this transient state, key signaling components relevant for neural induction and neural stem cell maintenance are regulated by and functionally contribute to iN reprogramming and maturation. Thus, Ascl1- and Sox2-mediated reprogramming into a broad spectrum of iN types involves the unfolding of a developmental program via neural stem cell-like intermediates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
15 August 2018
In the version of this article initially published online, Supplementary Table 7 could not be opened. The error has been corrected online.
References
Heinrich, C., Spagnoli, F. M. & Berninger, B. In vivo reprogramming for tissue repair. Nat. Cell Biol. 17, 204–211 (2015).
Amamoto, R. & Arlotta, P. Development-inspired reprogramming of the mammalian central nervous system. Science 343, 1239882 (2014).
Mertens, J., Marchetto, M. C., Bardy, C. & Gage, F. H. Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat. Rev. Neurosci. 17, 424–437 (2016).
Srivastava, D. & DeWitt, N. In vivo cellular reprogramming: the next generation. Cell 166, 1386–1396 (2016).
Ruiz, S. et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 21, 45–52 (2011).
Morris, S. A. Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks. Development 143, 2696–2705 (2016).
Vierbuchen, T. & Wernig, M. Molecular roadblocks for cellular reprogramming. Mol. Cell 47, 827–838 (2012).
Gascón, S., Masserdotti, G., Russo, G. L. & Götz, M. Direct neuronal reprogramming: achievements, hurdles, and new roads to success. Cell Stem Cell 21, 18–34 (2017).
Wapinski, O. L. et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635 (2013).
Masserdotti, G. et al. Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell 17, 74–88 (2015).
Treutlein, B. et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395 (2016).
Mall, M. et al. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature 544, 245–249 (2017).
Heinrich, C. et al. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 8, e1000373 (2010).
Chanda, S., Marro, S., Wernig, M. & Südhof, T. C. Neurons generated by direct conversion of fibroblasts reproduce synaptic phenotype caused by autism-associated neuroligin-3 mutation. Proc. Natl. Acad. Sci. USA 110, 16622–16627 (2013).
Vierbuchen, T. et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010).
Karow, M. et al. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 11, 471–476 (2012).
Long, J. E. et al. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J. Comp. Neurol. 512, 556–572 (2009).
Liu, S. J. et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17, 67 (2016).
Raposo, A.A. et al. Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. https://doi.org/10.1016/j.celrep.2015.02.025 (2015).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Camp, J. G. et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 112, 15672–15677 (2015).
Suzuki, I. K. & Vanderhaeghen, P. Is this a brain which I see before me? Modeling human neural development with pluripotent stem cells. Development 142, 3138–3150 (2015).
Imayoshi, I., Sakamoto, M., Yamaguchi, M., Mori, K. & Kageyama, R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 30, 3489–3498 (2010).
Guillemot, F. Spatial and temporal specification of neural fates by transcription factor codes. Development 134, 3771–3780 (2007).
Ladewig, J. et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 9, 575–578 (2012).
Dias Moura Prazeres, P. H. et al. Pericytes are heterogeneous in their origin within the same tissue. Dev. Biol. 427, 6–11 (2017).
La Manno, G. et al. Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19 (2016).
Göritz, C. et al. A pericyte origin of spinal cord scar tissue. Science 333, 238–242 (2011).
Chambers, S. M. et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009).
Liu, M. L. et al. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4, 2183 (2013).
Kageyama, R., Ohtsuka, T., Shimojo, H. & Imayoshi, I. Dynamic regulation of Notch signaling in neural progenitor cells. Curr. Opin. Cell Biol. 21, 733–740 (2009).
Vasconcelos, F. F. et al. MyT1 counteracts the neural progenitor program to promote vertebrate neurogenesis. Cell Rep. 17, 469–483 (2016).
Engler, A. et al. Notch2 signaling maintains NSC quiescence in the murine ventricular-subventricular zone. Cell Rep. 22, 992–1002 (2018).
Radonjić, N. V. et al. Diversity of cortical interneurons in primates: the role of the dorsal proliferative niche. Cell Rep. 9, 2139–2151 (2014).
Karow, M., Schichor, C., Beckervordersandforth, R. & Berninger, B. Lineage-reprogramming of pericyte-derived cells of the adult human brain into induced neurons. J. Vis. Exp. https://doi.org/10.3791/51433 (2014).
Yu, P. B. et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 4, 33–41 (2008).
Inman, G. J. et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 62, 65–74 (2002).
Laping, N. J. et al. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol. Pharmacol. 62, 58–64 (2002).
Martynoga, B. et al. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 27, 1769–1786 (2013).
Graham, A., Francis-West, P., Brickell, P. & Lumsden, A. The signalling molecule BMP4 mediates apoptosis in the rhombencephalic neural crest. Nature 372, 684–686 (1994).
Chen, A. E., Borowiak, M., Sherwood, R. I., Kweudjeu, A. & Melton, D. A. Functional evaluation of ES cell-derived endodermal populations reveals differences between Nodal and Activin A-guided differentiation. Development 140, 675–686 (2013).
Kumar, A. et al. Nodal signaling uses activin and transforming growth factor-beta receptor-regulated Smads. J. Biol. Chem. 276, 656–661 (2001).
Schubert, W. Multiple antigen-mapping microscopy of human tissue. in Experta Medica: Advances in Analytical Cellular Patphology (eds. Burger, G., Oberholzer, M. & Vooijs, G.P.) 97–98 (Elsevier, Amsterdam, 1990).
Ferreira, T. A. et al. Neuronal morphometry directly from bitmap images. Nat. Methods 11, 982–984 (2014).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Zemankovics, R., Káli, S., Paulsen, O., Freund, T. F. & Hájos, N. Differences in subthreshold resonance of hippocampal pyramidal cells and interneurons: the role of h-current and passive membrane characteristics. J. Physiol. 588, 2109–2132 (2010).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Oshlack, A., Robinson, M. D. & Young, M. D. From RNA-seq reads to differential expression results. Genome Biol. 11, 220 (2010).
Alexa, A., Rahnenführer, J. & Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 (2006).
Renaud, G., Kircher, M., Stenzel, U. & Kelso, J. freeIbis: an efficient basecaller with calibrated quality scores for Illumina sequencers. Bioinformatics 29, 1208–1209 (2013).
Renaud, G., Stenzel, U., Maricic, T., Wiebe, V. & Kelso, J. deML: robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics 31, 770–772 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Acknowledgements
We thank M. Wernig (Stanford University) for generously providing us with the Sox2 coding sequence. We are also very grateful to B. Sutor (BMC, LMU Munich) for help with the electrophysiological experiments, R. Menon (UMC Mainz) for help with the cell culture experiments, A. Bosio (Miltenyi Biotec) for help with the multidimensional fluorescence stainings, and F. Calzolari (UMC Mainz) for comments on the manuscript. We thank B. Höber, A. Weihmann, and J. Kelso of MPI-EVA for sequencing and bioinformatics support with this project. Flow cytometric cell sorting was performed at the “Core Unit Durchflusszytometrie” (CUDZ) of the Center for Infectious Diseases at the College of Veterinary Medicine, University of Leipzig, Leipzig, Germany. S.F. was supported by a fellowship from the Swiss National Science Foundation (PA00P3_139709). W.F. was supported by a Fellowship from the China Research Council. This work was supported by the following grants: advanced ERC ChroNeuroRepair to M.G.; Bavarian State Ministry of Sciences, Research and the Arts to M.K. and B.B. (ForIPS D2-F2412.26); Schram foundation (T287/29577/2017) and Wings For Life (WFL-DE-012/14) to M.K.; Max Planck Society to B.T.; and DFG (INST 161/875-2; BE 4182/8-1), NEURON ERA-NET (01EW1604), Wellcome Trust (206410/Z/17/Z), and the research initiative “Wissen schafft Zukunft” of Rhineland-Palatinate to B.B.
Author information
Authors and Affiliations
Contributions
M.K., J.G.C., B.T., and B.B. conceived the study and designed experiments; M.K. performed direct reprogramming experiments; M.K., S.F., A.P., and V.K.T. analyzed bulk RNA-seq data; A.B. helped with processing of the 10xGenomics data; A.G. performed RNA isolation for bulk RNA-seq analysis; W.F. performed time-lapse imaging experiments; T.R. performed electrophysiological recordings; A.C. performed Sholl analyses; A.S. performed immunocytochemical analyses; C.S. provided human brain biopsies; M.G. provided material; J.G.C., M.G.-S., and T.G. performed single-cell RNA-seq experiments and sequenced libraries; J.G.C., J.K., and B.T. analyzed single-cell RNA-seq data; all authors discussed the data; and M.K., J.G.C., S.F., B.T., and B.B. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Cooperative AS function is required for pericyte-to-iN conversion.
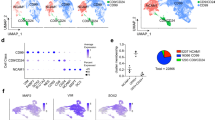
a, Euler diagram shows number of DE genes of each condition (Ascl1-only, Sox2-only, AS) at 2 dpi. b, Bar graph representing number of direct Ascl1-targets transcriptionally upregulated across different conditions and timepoints. c, Euler diagram showing that the majority of direct Ascl1-targets are regulated by AS synergism in human brain pericytes at 2 and 7 dpi. d, Heatmap shows normalized expression (Z-score) of direct Ascl1-target genes upregulated in AS-transduced cells at 7 dpi. Note that only a minor fraction of these genes is also upregulated by Ascl1 alone. e, GO term analysis of DE genes for each condition at 2 dpi. Shown are the 10 most significantly regulated GO terms. GO terms were ordered according to their significance as determined by Fisher´s exact test; n = 3 individual pericyte donors per experimental group. f, GO term analysis of DE genes for each condition at 7 dpi. Significantly regulated genes in GO categories are contained in Supplementary Table 2. GO terms were ordered according to their significance as determined by Fisher´s exact test; n = 3 individual pericyte donors per experimental group. g, tSNE plots from the analysis of Ascl1-only and AS transduced cells at 2 and 7 dpi from Fig. 1e are colored based on the expression of selected pericyte genes (Log2 FPKM). h, tSNE plots from the analysis of Ascl1-only and AS transduced cells at 2 and 7 dpi from Fig. 1e are colored based on the expression of selected mesoderm, neurogenesis-related, and GABAergic signature genes (Log2 FPKM).
Supplementary Figure 2 Expression of pericyte heterogeneity genes.
a, tSNE plot from Fig. 2a colored based on the expression of selected genes enriched in different pericyte groups. b, Violin plots show the density expression distribution of genes in pericyte group 1 (31 cells) and group 2 (44 cells). c, GO term analysis of pericyte heterogeneity genes from Fig. 2b; group 1, n = 31 cells; group 2, n = 44 cells; GO terms were ordered according to their significance as determined by Fisher´s exact test. d, Representative flow cytometry plot showing isotype control for the sorting of LEPR-positive cultured human pericytes (Fig. 2f); n = 4.
Supplementary Figure 3 Switch genes are expressed in the germinal zones of the developing CNS.
a In situ hybridization images (GenePaint.org) show the expression of selected switch genes in the developing mouse forebrain (E14.5). b Projecting the switch gene signature onto published scRNA-seq data1 reconstructing the MEF-to-iN reprogramming path (259 cells) shows the high base level in the starting MEF population and downregulation of the switch gene signature along differentiation towards iNs. Shaded gray represents 0.95 confidence interval.
Supplementary Figure 4 Characterization of ASD cells and shift towards a glutamatergic neuron phenotye by combinatorial expression of AS with Neurog2.
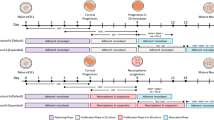
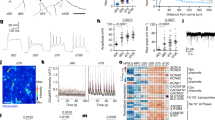
a, Summary of morphometric analyses showing significant differences between untreated and Dorsomorphin-treated AS cells. Data are shown as mean ± SEM; (untreated, n = 14 cells of 3 independent experiments; Dorsomorphin-treated, n = 14 cells of 3 independent experiments); two-tailed unpaired Student's t test; primary branches, P = 0.015; number of dendritic segments, P = 0.0005; branching points, P = 0.001; maximum length, P = 0.035; soma size, P = 0.0006; sum of intersections P = 0.02; *P < 0.05, ** P < 0.01, ***P < 0.001. b, Electrophysiological assessment of AS and ASD cells. Representative traces of multiple action potential discharge after step-current injections in AS (left) and ASD (middle) cells. Action potentials could be reliably blocked by TTX [0.5 µM] bath application (right). c, Micrographs show iGNs treated with Dorsomorphin immunoreactive for TUBB3 and GABA which acquire highly complex neuronal morphology. ASD iGNs show overlapping immunoreactivity for parvalbumin (PVALB) and GABA (n =3). Nuclei are stained with Dapi. Scale bars = 50 µm. d, Biplot showing the expression of DLX1 and NEUROG2 in all SNAP25-expressing cells. e, Monocle plot from Fig. 4g colored based on the expression of genes that show the loss of pericyte marker gene expression along the pseudotime and the acquisition of GABAergic and glutamatergic cell fate determinants along the trajectory towards different neuronal subtypes. f, Schematic of ASN experiments. g, Micrographs show pericytes transduced with AS (green) and Neurog2 (red) that acquire VGLUT1 immunoreactivity by co-expression of these three transcription factors. Note the punctate appearance of VGLUT1 in inset (right) (n = 3). Nuclei are stained with Dapi. Scale bars = 50 µm.
Supplementary Figure 5 High-throughput scRNA-seq data from a second pericyte donor confirms lineage bifurcations during iN maturation.
a, scRNA-seq using a high-throughput droplet microfluidic platform (10X genomics) was performed on 3419 AS-transduced cells treated with Dorsomorphin at 14 days post infection. PCA followed by tSNE shows cell populations that maintain pericyte markers (greys) and fail to productively differentiate to a neuronal lineage, populations at intermediate stages of differentiation (light grey), and two distinct neuron populations (cyan, blue). b, Cells are colored in the tSNE plots based on log normalized expression of pericyte marker PDGFRB, group 1 marker LEPR, and neuronal marker SNAP25. c, The inset shows the log normalized expression of markers for distinct excitatory (NEUROG2) and inhibitory (DLX1) neuronal populations that emerge during reprogramming. d, Heatmap shows the scaled expression of the top 20 genes that are differentially expressed (based on average log fold change) between the two neuronal populations. Single cells are in columns, genes in rows.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5
Supplementary Table 1
DE analysis of bulk RNA-sequencing data following overexpression of Ascl1, Sox2, Ascl1-Sox2, and control vector.
Supplementary Table 2
GO terms including significant genes within GO terms of DE genes from bulk RNA-seq analysis.
Supplementary Table 3
Comparison of DE genes with direct Ascl1-targets.
Supplementary Table 4
Genes in GO terms of pericyte group 1 and group 2.
Supplementary Table 5
Fluidigm C1 transcriptome data for all 769 cells with annotations (quantification in log2[FPKM]).
Supplementary Table 6
Genes used for calculating pericyte, mesodermal, neurogenesis-related, and GABAergic signatures.
Supplementary Table 7
10x Genomics transcriptome data for all 3419 sorted ASD cells.
Supplementary Video 1
Time-lapse video microscopy of human brain pericytes transduced with AS and treated with Dorsomorphin.
Rights and permissions
About this article
Cite this article
Karow, M., Camp, J.G., Falk, S. et al. Direct pericyte-to-neuron reprogramming via unfolding of a neural stem cell-like program. Nat Neurosci 21, 932–940 (2018). https://doi.org/10.1038/s41593-018-0168-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-018-0168-3
This article is cited by
-
Potential use of iPSCs for disease modeling, drug screening, and cell-based therapy for Alzheimer’s disease
Cellular & Molecular Biology Letters (2023)
-
Efficient generation of functional neurons from mouse embryonic stem cells via neurogenin-2 expression
Nature Protocols (2023)
-
Transfer learning enables predictions in network biology
Nature (2023)
-
Targeting Pericytes for Functional Recovery in Ischemic Stroke
NeuroMolecular Medicine (2023)
-
ASCL1 activates neuronal stem cell-like lineage programming through remodeling of the chromatin landscape in prostate cancer
Nature Communications (2022)