Abstract
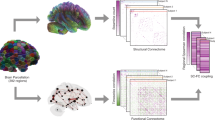
Microscopic features (that is, microstructure) of axons affect neural circuit activity through characteristics such as conduction speed. To what extent axonal microstructure in white matter relates to functional connectivity (synchrony) between brain regions is largely unknown. Using MRI data in 11,354 subjects, we constructed multivariate models that predict functional connectivity of pairs of brain regions from the microstructural signature of white matter pathways that connect them. Microstructure-derived models provided predictions of functional connectivity that explained 3.5% of cross-subject variance on average (ranging from 1–13%, or r = 0.1–0.36) and reached statistical significance in 90% of the brain regions considered. The microstructure–function relationships were associated with genetic variants, co-located with genes DAAM1 and LPAR1, that have previously been linked to neural development. Our results demonstrate that variation in white matter microstructure predicts a fraction of functional connectivity across individuals, and that this relationship is underpinned by genetic variability in certain brain areas.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All source data (including raw and processed brain imaging data and genetics data) are available from UK Biobank via their standard data access procedure (see http://www.ukbiobank.ac.uk/register-apply).
Code availability
The image processing pipelines of the MRI data in the UK Biobank project can be found at http://www.fmrib.ox.ac.uk/ukbiobank. Custom-written Matlab code including the microstructure–function modeling is freely available at https://users.fmrib.ox.ac.uk/~jmollink/Biobank/Biobank.html.
References
Buzsáki, G. Rhythms of the Brain (Oxford University Press, 2006).
Kötter, R. & Sommer, F. T. Global relationship between anatomical connectivity and activity propagation in the cerebral cortex. Phil. Trans. R. Soc. Lond. B 355, 127–134 (2000).
Rorden, C. & Karnath, H.-O. Using human brain lesions to infer function: a relic from a past era in the fMRI age? Nat. Rev. Neurosci. 5, 813–819 (2004).
Schmahmann, J. D. & Pandya, D. N. in Fiber Pathways of the Brain 1–37 (Oxford University Press, 2006).
Basser, P. J., Pajevic, S., Pierpaoli, C., Duda, J. & Aldroubi, A. In vivo fiber tractography using DT-MRI data. Magn. Reson. Med. 44, 625–632 (2000).
Jbabdi, S. & Johansen-Berg, H. Tractography: where do we go from here? Brain Connect. 1, 169–183 (2011).
Miller, K. L. et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19, 1523–1536 (2016).
Greicius, M. D., Supekar, K., Menon, V. & Dougherty, R. F. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb. Cortex 19, 72–78 (2009).
van den Heuvel, M. P., Mandl, R. C. W., Kahn, R. S. & Hulshoff Pol, H. E. Functionally linked resting-state networks reflect the underlying structural connectivity architecture of the human brain. Hum. Brain Mapp. 30, 3127–3141 (2009).
Hagmann, P. et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 6, e159 (2008).
Koch, M. A., Norris, D. G. & Hund-Georgiadis, M. An investigation of functional and anatomical connectivity using magnetic resonance imaging. Neuroimage 16, 241–250 (2002).
Beaulieu, C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 15, 435–455 (2002).
van den Heuvel, M., Mandl, R., Luigjes, J. & Hulshoff Pol, H. Microstructural organization of the cingulum tract and the level of default mode functional connectivity. J. Neurosci. 28, 10844–10851 (2008).
Wahl, M. et al. Human motor corpus callosum: topography, somatotopy, and link between microstructure and function. J. Neurosci. 27, 12132–12138 (2007).
Zhang, H., Schneider, T., Wheeler-Kingshott, C. A. & Alexander, D. C. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016 (2012).
Elliott, L. T. et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216 (2018).
Stark, D. E. et al. Regional variation in interhemispheric coordination of intrinsic hemodynamic fluctuations. J. Neurosci. 28, 13754–13764 (2008).
Shen, K. et al. Stable long-range interhemispheric coordination is supported by direct anatomical projections. Proc. Natl Acad. Sci. USA 112, 6473–6478 (2015).
Alfaro-Almagro, F. et al. Image processing and quality control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 166, 400–424 (2018).
Basser, P. J., Mattiello, J. & LeBihan, D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. 103, 247–254 (1994).
Ennis, D. B. & Kindlmann, G. Orthogonal tensor invariants and the analysis of diffusion tensor magnetic resonance images. Magn. Reson. Med. 55, 136–146 (2006).
Behrens, T. E. J., Berg, H. J., Jbabdi, S., Rushworth, M. F. S. & Woolrich, M. W. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage 34, 144–155 (2007).
Smith, S. M. et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 31, 1487–1505 (2006).
Mollink, J. et al. Evaluating fibre orientation dispersion in white matter: comparison of diffusion MRI, histology and polarized light imaging. Neuroimage 157, 561–574 (2017).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Matusek, T. et al. Formin proteins of the DAAM subfamily play a role during axon growth. J. Neurosci. 28, 13310–13319 (2008).
Avilés, E. C. & Stoeckli, E. T. Canonical wnt signaling is required for commissural axon guidance. Dev. Neurobiol. 76, 190–208 (2016).
Won, H. et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016).
Wang, Y. et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19, 151 (2018).
Yung, Y. C., Stoddard, N. C., Mirendil, H. & Chun, J. Lysophosphatidic acid signaling in the nervous system. Neuron 85, 669–682 (2015).
Hibar, D. P. et al. Common genetic variants influence human subcortical brain structures. Nature 520, 224–229 (2015).
Honey, C. J. et al. Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl Acad. Sci. USA 106, 2035–2040 (2009).
O’Reilly, J. X. et al. Causal effect of disconnection lesions on interhemispheric functional connectivity in rhesus monkeys. Proc. Natl Acad. Sci. USA 110, 13982–13987 (2013).
Roland, J. L. et al. On the role of the corpus callosum in interhemispheric functional connectivity in humans. Proc. Natl Acad. Sci. USA 114, 13278–13283 (2017).
Smith, S. M. & Nichols, T. E. Statistical challenges in “big data” human neuroimaging. Neuron 97, 263–268 (2018).
Torkamani, A., Wineinger, N. E. & Topol, E. J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018).
Tobyne, S. M. et al. A surface-based technique for mapping homotopic interhemispheric connectivity:development, characterization, and clinical application. Hum. Brain Mapp. 37, 2849–2868 (2016).
Schmahmann, J. D. et al. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain 130, 630–653 (2007).
Smith, S. M. The future of FMRI connectivity. Neuroimage 62, 1257–1266 (2012).
Cordes, D. et al. Frequencies contributing to functional connectivity in the cerebral cortex in “resting-state” data. Am. J. Neuroradiol. 22, 1326–1333 (2001).
Friston, K. J. Functional and effective connectivity: a review. Brain Connect. 1, 13–36 (2011).
Budde, M. D. & Annese, J. Quantification of anisotropy and fiber orientation in human brain histological sections. Front. Integr. Neurosci. 7, 3 (2013).
Hibar, D. P. et al. Novel genetic loci associated with hippocampal volume. Nat. Commun. 8, 13624 (2017).
Habas, R., Kato, Y. & He, X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107, 843–854 (2001).
Salomon, S. N., Haber, M., Murai, K. K. & Dunn, R. J. Localization of the Diaphanous-related formin Daam1 to neuronal dendrites. Neurosci. Lett. 447, 62–67 (2008).
Kida, Y., Shiraishi, T. & Ogura, T. Identification of chick and mouse Daam1 and Daam2 genes and their expression patterns in the central nervous system. Brain Res. Dev. Brain Res. 153, 143–150 (2004).
Behrends, U. et al. Novel tumor antigens identified by autologous antibody screening of childhood medulloblastoma cDNA libraries. Int. J. Cancer 106, 244–251 (2003).
González de San Román, E. et al. Anatomical location of LPA1 activation and LPA phospholipid precursors in rodent and human brain. J. Neurochem. 134, 471–485 (2015).
Fields, R. D. A new mechanism of nervous system plasticity: activity-dependent myelination. Nat. Rev. Neurosci. 16, 756–767 (2015).
Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W. & Smith, S. M. FSL. Neuroimage 62, 782–790 (2012).
Jenkinson, M., Bannister, P., Brady, M. & Smith, S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002).
Salimi-Khorshidi, G. et al. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage 90, 449–468 (2014).
Andersson, J. L. R. & Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 125, 1063–1078 (2016).
Daducci, A. et al. Accelerated microstructure imaging via convex pptimization (AMICO) from diffusion MRI data. Neuroimage 105, 32–44 (2015).
Griffanti, L. et al. BIANCA (Brain Intensity AbNormality Classification Algorithm): a new tool for automated segmentation of white matter hyperintensities. Neuroimage 141, 191–205 (2016).
Beckmann, C. F. & Smith, S. M. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging 23, 137–152 (2004).
Behrens, T. E. J. et al. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn. Reson. Med. 50, 1077–1088 (2003).
Jbabdi, S., Sotiropoulos, S. N., Savio, A. M., Graña, M. & Behrens, T. E. J. Model-based analysis of multishell diffusion MR data for tractography: how to get over fitting problems. Magn. Reson. Med. 68, 1846–1855 (2012).
de Groot, M. et al. Improving alignment in tract-based spatial statistics: evaluation and optimization of image registration. Neuroimage 76, 400–411 (2013).
Bach, M. et al. Methodological considerations on tract-based spatial statistics (TBSS). Neuroimage 100, 358–369 (2014).
Winkler, A. M., Ridgway, G. R., Webster, M. A., Smith, S. M. & Nichols, T. E. Permutation inference for the general linear model. Neuroimage 92, 381–397 (2014).
Sotiropoulos, S. N., Behrens, T. E. J. & Jbabdi, S. Ball and rackets: inferring fiber fanning from diffusion-weighted MRI. Neuroimage 60, 1412–1425 (2012).
Budde, M. D. & Frank, J. A. Examining brain microstructure using structure tensor analysis of histological sections. Neuroimage 63, 1–10 (2012).
Heinrich, M. P. et al. MIND: modality independent neighbourhood descriptor for multi-modal deformable registration. Med. Image Anal. 16, 1423–1435 (2012).
Acknowledgements
All data in this study were obtained from the UK Biobank project (access no. 8107). We are very grateful to all individuals who donated their time to participate in the UK Biobank study. K.L.M., M.K. and J.Mo. are supported by the Wellcome Trust (nos. 091509/Z/10/Z, 202788/Z/16/Z, 098369/Z/12/Z). The authors gratefully acknowledge funding from the Wellcome Trust UK Strategic Award (no. 098369/Z/12/Z). UK Biobank brain imaging and F.A.-A. are funded by the UK Medical Research Council and the Wellcome Trust. J.Ma. acknowledges funding for this work from the European Research Council (grant no. 617306) and the Leverhulme Trust. S.J. is supported by the UK Medical Research Council (no. MR/L009013/1). The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (no. 203139/Z/16/Z). Finally, we would like to thank D. Norris and J. Marques for their valuable input on this work.
Author information
Authors and Affiliations
Contributions
J.Mo., S.M.S., S.J. and K.L.M. designed the research. J.Mo. performed the research. K.L.M., F.A.-A. and S.M.S., developed acquisition and processing pipelines for the MRI data. L.T.E. and J.Ma. processed genetics data, provided tools for genome-wide associations analysis and gave feedback on genetics results. J.Mo., S.M.S., M.K., M.H., A.M.C.W., S.J. and K.L.M. analyzed the data and interpreted its outcomes. J.Mo. and K.L.M. wrote the manuscript, which was edited by all authors.
Corresponding author
Ethics declarations
Competing interests
J.Ma. is a co-founder and director of GENSCI Ltd. S.S. is a co-founder of SBGneuro.
Additional information
Journal peer review information: Nature Neuroscience thanks Genevieve Konopka and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mollink, J., Smith, S.M., Elliott, L.T. et al. The spatial correspondence and genetic influence of interhemispheric connectivity with white matter microstructure. Nat Neurosci 22, 809–819 (2019). https://doi.org/10.1038/s41593-019-0379-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-019-0379-2
This article is cited by
-
Impact of corpus callosum integrity on functional interhemispheric connectivity and cognition in healthy subjects
Brain Imaging and Behavior (2023)
-
Genomics of perivascular space burden unravels early mechanisms of cerebral small vessel disease
Nature Medicine (2023)
-
The interindividual variability of multimodal brain connectivity maintains spatial heterogeneity and relates to tissue microstructure
Communications Biology (2022)
-
Corticospinal excitability and conductivity are related to the anatomy of the corticospinal tract
Brain Structure and Function (2022)
-
Functional cortical associations and their intraclass correlations and heritability as revealed by the fMRI Human Connectome Project
Experimental Brain Research (2022)