Abstract
While CNS microglia have been extensively studied, relatively little is known about macrophages populating the peripheral nervous system. Here we performed ontogenic, transcriptomic and spatial characterization of sciatic nerve macrophages (snMacs). Using multiple fate-mapping systems, we show that snMacs do not derive from the early embryonic precursors colonizing the CNS, but originate primarily from late embryonic precursors and become replaced by bone-marrow-derived macrophages over time. Using single-cell transcriptomics, we identified a tissue-specific core signature of snMacs and two spatially separated snMacs: Relmα+Mgl1+ snMacs in the epineurium and Relmα–Mgl1– snMacs in the endoneurium. Globally, snMacs lack most of the core signature genes of microglia, with only the endoneurial subset expressing a restricted number of these genes. In response to nerve injury, the two resident snMac populations respond differently. Moreover, and unlike in the CNS, monocyte-derived macrophages that develop during injury can engraft efficiently in the pool of resident peripheral nervous system macrophages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Raw data may be available upon reasonable request to the corresponding authors. The raw RNA-seq data were uploaded to the NCBI GEO under the accession code GSE144708.
Code availability
The scripts used for the gene expression analysis can be found at https://github.com/saeyslab/MFs_peripheralCentralNerveSystem
References
Wynn, T. A., Chawla, A. & Pollard, J. W. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013).
Scott, C. L. et al. The transcription factor ZEB2 is required to maintain the tissue-specific identities of macrophages. Immunity 49, 312–325.e5 (2018).
Haimon, Z. et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 19, 636–644 (2018).
Mildner, A. et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat. Neurosci. 10, 1544–1553 (2007).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Ajami, B., Bennett, J. L., Krieger, C., Tetzlaff, W. & Rossi, F. M. V. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10, 1538–1543 (2007).
Jordão, M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554 (2019).
Van Hove, H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22, 1021–1035 (2019).
Ajami, B., Bennett, J. L., Krieger, C., McNagny, K. M. & Rossi, F. M. V. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci. 14, 1142–1149 (2011).
Mass, E. et al. Specification of tissue-resident macrophages during organogenesis. Science 353, aaf4238 (2016).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Kierdorf, K. et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013).
Chen, P., Piao, X. & Bonaldo, P. Role of macrophages in Wallerian degeneration and axonal regeneration after peripheral nerve injury. Acta Neuropathol. 130, 605–618 (2015).
Cattin, A.-L. et al. Macrophage-induced blood vessels guide Schwann cell-mediated regeneration of peripheral nerves. Cell 162, 1127–1139 (2015).
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Guilliams, M. & Scott, C. L. Does niche competition determine the origin of tissue-resident macrophages? Nat. Immunol. 17, 451–460 (2017).
Scott, C. L. et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016).
van den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017).
Saunders, A. et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16 (2018).
Mrdjen, D. et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395.e6 (2018).
Goldmann, T. et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805 (2016).
Matcovitch-Natan, O. et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670 (2016).
Ginhoux, F. & Prinz, M. Origin of microglia: current concepts and past controversies. Cold Spring Harb. Perspect. Biol. 7, a020537 (2015).
Prinz, M., Erny, D. & Hagemeyer, N. Ontogeny and homeostasis of CNS myeloid cells. Nat. Immunol. 18, 385–392 (2017).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Werner, Y. et al. Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke. Nat. Neurosci. 23, 351–362 (2020).
Hoeffel, G. et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015).
Serbina, N. V. & Pamer, E. G. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 7, 311–317 (2006).
Movahedi, K. et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 70, 5728–5739 (2010).
Nadeau, S. et al. Functional recovery after peripheral nerve injury is dependent on the pro-inflammatory cytokines IL-1β and TNF: implications for neuropathic pain. J. Neurosci. 31, 12533–12542 (2011).
Shechter, R. et al. Recruitment of beneficial M2 macrophages to injured spinal cord is orchestrated by remote brain choroid plexus. Immunity 38, 555–569 (2013).
Jablonski, K. A. et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS ONE 10, e0145342 (2015).
Chakarov, S. et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019).
Tamoutounour, S. et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938 (2013).
Wang, P. L. et al. Peripheral nerve resident macrophages are microglia-like cells with tissue-specific programming. Preprint at biorXiv https://www.biorxiv.org/content/10.1101/2019.12.19.883546v1 (2019).
Shemer, A. et al. Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat. Commun. 9, 5206 (2018).
Bennett, F. C. et al. A combination of ontogeny and CNS environment establishes microglial identity. Neuron 98, 1170–1183.e8 (2018).
Cronk, J. C. et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med. 215, 1627–1647 (2018).
Buttgereit, A. et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 17, 1397–1406 (2016).
van de Laar, L. et al. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44, 755–768 (2016).
Bonnardel, J. et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity 51, 638–654.e9 (2019).
Ydens, E. et al. Acute injury in the peripheral nervous system triggers an alternative macrophage response. J. Neuroinflammation 9, 176 (2012).
Meisel, C., Schwab, J. M., Prass, K., Meisel, A. & Dirnagl, U. Central nervous system injury-induced immune deficiency syndrome. Nat. Rev. Neurosci. 6, 775–786 (2005).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017).
Inoue, S., Inoue, M., Fujimura, S. & Nishinakamura, R. A mouse line expressing Sall1-driven inducible Cre recombinase in the kidney mesenchyme. Genesis 48, 207–212 (2010).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4–8 (2001).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Guilliams, M. et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210, 1977–1992 (2013).
Ydens, E. et al. Nlrp6 promotes recovery after peripheral nerve injury independently of inflammasomes. J. Neuroinflammation 12, 629 (2015).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Schindelin, J., Rueden, C. T., Hiner, M. C. & Eliceiri, K. W. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 (2015).
Lun, A. T. L., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122 (2016).
Acknowledgements
We would like to thank G. Van Isterdael from the Flow Cytometry Core Facility for assistance. This study benefitted from data assembled by the ImmGen Consortium. We would also like to thank VIB TechWatch and the VIB Single Cell Accelerator program for supporting the 10× Genomics experiments. Y.S.’s Lab is supported by the FWO, and Y.S. is a Marylou Ingram Scholar. C.L.S. is supported by FWO grants. M.G.’s Lab is supported by grants from the FWO (3G069219 FWO grant on nerve macrophages), an ERC Consolidator grant (725924) and a single-cell technology Ghent University GOA grant. M.P. is supported by the Sobek Foundation, the Ernst-Jung Foundation, the German Research Foundation (SFB 992, SFB1160, DFG SFB/TRR167, Reinhart-Koselleck-Grant, Gottfried Wilhelm Leibniz-Prize) and the Ministry of Science, Research and Arts, Baden-Wuerttemberg (Sonderlinie “Neuroinflammation”) and by the BMBF-funded competence network of multiple sclerosis (KKNMS). His research is supported by the German Research Foundation (DFG) under Germany’s Excellence Strategy (CIBSS–EXC-2189–Project ID 390939984). F.G. is an EMBO YIP awardee and is supported by Singapore Immunology Network (SIgN) core funding as well as a Singapore National Research Foundation Senior Investigatorship (NRFI) NRF2016NRF-NRFI001-02. The study was supported by the University of Antwerp and the Fund for Scientific research (FWO-Flanders). E.Y. was supported by an FWO-SB doctoral fellowship. S.J. is a recipient of an ERC Consolidator Grant (819314), several FWO program grants and an EOS grant (G0G7318N).
Author information
Authors and Affiliations
Contributions
E.Y., L.A., F.G., M.P., S.J. and M.G. conceived the project. E.Y., L.A., B.A., C.L.S., D.S., O.M., T.B., S.D.P., D.L. and T.M. performed the experiments. L.M., E.Y. and L.A. analyzed the data. E.Y., L.A., M.P., S.J. and M.G. designed the figures. Y.S., V.T., F.G., S.J., M.P. and M.G. supervised the experiments and data analyses. R.S. provided the Cxcr4CreERT2 mouse line. E.Y., L.A., M.P., S.J. and M.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Ozgun Gokce, Aakanksha Jain, Clifford Woolf, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
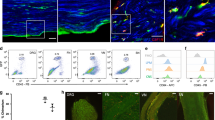
Extended Data Fig. 1 Gating strategy for sorting snMacs, onMacs and brain microglia for bulk RNA sequencing.
Top, representative gating strategy used to sort CD64+ snMacs (top) and purity check (down). The sciatic nerve from at least 5 mice were pooled and stained for the displayed surface markers. CD64+ cells among CD45+ cells were removed of doublets, dead cells, eosinophils, neutrophils and monocytes to obtain CD11bInt-HiCD64Hi snMacs. Bottom, representative gating strategies used to sort CD64+F4/80+ snMacs macrophages (top), CD45LoCD11bHi onMacs (middle) and CD45LoCD11bHi brain microglia (bottom) are displayed. The sciatic nerves and optic nerves were harvested from the same mouse and nerves of five individual animals were pooled for one biological replicate. Four individual biological replicates from independent experiments were submitted to RNA sequencing. For brain microglia, cells of five individual mice were sorted and submitted to RNA sequencing. Dump channel included antibodies against Ly6C, Ly6G, CD3e, CD19 and B220.
Extended Data Fig. 2 Most genes identified by bulk transcriptomics are expressed by either epineurial and endoneurial snMacs.
a, Single-cell heatmap of the genes identified in Figure 1 by bulk transcriptomics plotted on the tSNE populations identified by sc-RNA-sequencing as Relmα+Mgl1+ snMacs and Relmα-Mgl1- snMacs. The heatmap shows the scaled log2 normalised expression values. The log2 normalised expression values were scaled using the ‘scale_quantile’ function of the SCORPIUS package with default parameters (v1.0). b, tSNE plot depicting the expression of Cx3cr1 in snMacs c, Representative flow cytometry plots showing Cx3cr1gfp expression in Mgl1- snMacs (red) and Mgl1+ snMacs (blue). Data represents five pooled mice. d, Quantification of Cx3cr1-GFP+ Mgl1- snMacs and Mgl1+ snMacs. Data are shown as mean ±SEM. Symbols represent individual animals (p14:3 mice, p56: 4mice). e, Representative confocal microscopy of sciatic nerve vibratome cross sections from Cx3cr1gfp/wt mice. Location of anti-GFP+MGL1- snMacs (filled arrowheads) and anti-GFP-MGL1+ snMacs (open arrowheads) is depicted. Nuclei were counterstained with DAPI. Scale bar: overview 100µm, insets 20µm. Images are representative of three individual mice.
Extended Data Fig. 3 Endoneurial snMacs express higher levels of immediate early response genes.
a, Single-cell heatmap of the immediate early response genes induced by tissues digestions identified by Van Hove et al. and projected on the tSNE populations identified by sc-RNA-sequencing as Relmα+Mgl1+ snMacs and Relmα-Mgl1- snMacs. The heatmap shows the scaled log2 normalised expression values. The log2 normalised expression values were scaled using the ‘scale_quantile’ function of the SCORPIUS package with default parameters (v1.0). b, tSNE plot depicting the expression of canonical immediate early response genes on Relmα+Mgl1+ snMacs and Relmα-Mgl1- snMacs.
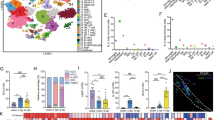
Extended Data Fig. 4 Heterogeneity of CNS and PNS macrophages.
a, tSNE plot containing the brain microglia and BAM populations described by Van Hove et al.10, as well as our snMacs. CPhi-BAM: MHCIIHi macrophages from the choroid plexus. CPlo-BAM: MHCIILow macrophages from the choroid plexus. Dhi-BAM: MHCIIHi macrophages from the Dura mater. Dlo-BAM: MHCIILow macrophages from the Dura mater. SD-BAM: macrophages from the Subdural meninges. Epineural snMacs: Relmα+Mgl1+ snMacs. Endoneurial snMacs: Relmα-Mgl1- snMacs. b, Feature plots of various CNS and PNS Mac-associated genes.
Extended Data Fig. 5 Mgl1+ epineurial macrophages express Lyve1 and are often located in close contact with blood vessels.
a, Expression of Lyve1 on snMacs subsets measured by flow cytometry. b, Representative histograms showing the expression of Lyve1 on Mgl1- endoneurial snMacs (orange) and Mgl1+ epineurial snMacs (blue) over FMO controls (grey). (a,b), Data are representative of five individual animals. Cells from all individual mice were pooled after recording in one plot. c, Confocal microscopy was used to verify the location of Lyve+ cells within the epineurium of the sciatic nerve. Insets depict Lyve1+ cells in close proximity to CD31+Collagen IV+ blood vessels (filled arrowheads). Not all Lyve1+ cells are found to be associated with the blood vessels (open arrowheads). d, F4/80+Lyve- endoneurial snMacs (open arrowheads) and F4/80+Lyve1+ epineurial snMacs (filled arrowheads) are depicted. Note that single F4/80+ epineurial snMacs were found to be Lyve1- (see R2). (c,d), The perineurium and the basal membranes of blood vessels were stained with Collagen IV. Nuclei were counterstained with DAPI. Images are representative of three individual mice.
Extended Data Fig. 6 Monocyte recruitment upon sciatic nerve injury.
a, We defined a large gate, called the “Mono-Mac waterfall”, containing Ly6CHi monocytes and CD64Hi macrophages among CD45+CD11b+Ly6G-SiglecF- cells in the sciatic nerve. The cells within the Mono-Mac waterfall gate (green), were then divided based on their Ly6C and MHCII expression. At day 0.5, incoming monocytes (red) can still be distinguished from resident macrophages (blue), even in the MHCIILow gate where the monocytes start to lose the Ly6C expression but still have lower CD64 expression. Once after Day 3, the Mono-Mac waterfall becomes one big continuum between Ly6CHi cells and CD64Hi cells. b, Number of macrophages (as defined by the Mono-Mac waterfall in (A)), Ly6G+Ly6C+ neutrophils, SiglecF+ eosinophils, CD3+MHCII- T cells and CD19+MHCII+ B cells in the sciatic nerve at the indicated timepoints after the injury. Data shown as mean ±SEM from a representative experiment with 3 mice per time-point, except for day 37 (2 mice). Two independent kinetic experiments were performed in total.
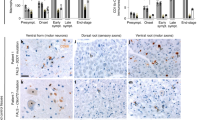
Extended Data Fig. 7 Relmα+ macrophage populations maintain their localization outside the nerve fascicles upon sciatic nerve injury and recovery.
Confocal microscopy was performed on nerve cross sections isolated from mice at different time points (0, 0.5, 1.5, 5 and 14 days) after nerve crush. Upon injury, macrophages (marked by F4/80) accumulate both inside the nerve fascicles and in the surrounding connective tissues. MHCIIHi and MHCIILow macrophage subpopulations do not harbor a specific localization and seem randomly distributed in different regions of the nerve. In contrast, at all examined time points Relmα+ cells were exclusively present in perineurium and epineurium connective tissues outside the nerve fascicles. Nerves from the different time points were collected from three separate nerve crush experiments (0.5 and 1.5 days; 0 and 5 days; 14 days) and images at each time point are representative of at least three individual mice. Scale bar = 100µm.
Supplementary information
Rights and permissions
About this article
Cite this article
Ydens, E., Amann, L., Asselbergh, B. et al. Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury. Nat Neurosci 23, 676–689 (2020). https://doi.org/10.1038/s41593-020-0618-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-0618-6
This article is cited by
-
Roles of Macrophages and Their Interactions with Schwann Cells After Peripheral Nerve Injury
Cellular and Molecular Neurobiology (2024)
-
Deletion of CD47 from Schwann cells and macrophages hastens myelin disruption/dismantling and scavenging in Schwann cells and augments myelin debris phagocytosis in macrophages
Journal of Neuroinflammation (2023)
-
M2 macrophage-derived cathepsin S promotes peripheral nerve regeneration via fibroblast–Schwann cell-signaling relay
Journal of Neuroinflammation (2023)
-
Pancreatic ductal adenocarcinoma induces neural injury that promotes a transcriptomic and functional repair signature by peripheral neuroglia
Oncogene (2023)
-
Microglia out of place—mapping macrophages across the developing human body
Cellular & Molecular Immunology (2023)