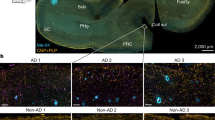
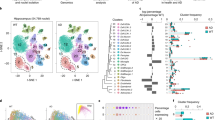
Abstract
Alzheimer’s disease (AD) is a complex neurodegenerative disease, perturbing neuronal and non-neuronal cell populations. In this study, using single-cell transcriptomics, we mapped all non-immune, non-neuronal cell populations in wild-type and AD model (5xFAD) mouse brains. We identified an oligodendrocyte state that increased in association with brain pathology, which we termed disease-associated oligodendrocytes (DOLs). In a murine model of amyloidosis, DOLs appear long after plaque accumulation, and amyloid-beta (Aβ) alone was not sufficient to induce the DOL signature in vitro. DOLs could be identified in a mouse model of tauopathy and in other murine neurodegenerative and autoimmune inflammatory conditions, suggesting a common response to severe pathological conditions. Using quantitative spatial analysis of mouse and postmortem human brain tissues, we found that oligodendrocytes expressing a key DOL marker (SERPINA3N/SERPINA3 accordingly) are present in the cortex in areas of brain damage and are enriched near Aβ plaques. In postmortem human brain tissue, the expression level of this marker correlated with cognitive decline. Altogether, this study uncovers a shared signature of oligodendrocytes in central nervous system pathologies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Raw and processed mouse sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus database under accession number GSE202297.
Code availability
All the code is available on GitHub at: https://github.com/PierreBSC/DOL_project
Change history
31 January 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41593-023-01266-y
References
Masters, C. L. et al. Alzheimer’s disease. Nat. Rev. Dis. Prim. 1, 15056 (2015).
Holtzman, D. M., Morris, J. C. & Goate, A. M. Alzheimer’s disease: the challenge of the second century. Sci. Transl. Med. 3, 77sr1 (2011).
Reitz, C. & Mayeux, R. Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem. Pharmacol. 88, 640–651 (2014).
Karch, C. M. & Goate, A. M. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 77, 43–51 (2015).
Schwartz, M. & Deczkowska, A. Neurological disease as a failure of brain–immune crosstalk: the multiple faces of neuroinflammation. Trends Immunol. 37, 668–679 (2016).
Heneka, M. T. et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14, 388–405 (2015).
Wyss-Coray, T. & Rogers, J. Inflammation in Alzheimer disease—a brief review of the basic science and clinical literature. Cold Spring Harb. Perspect. Med. 2, a006346 (2012).
De Strooper, B. & Karran, E. The cellular phase of Alzheimer’s disease. Cell 164, 603–615 (2016).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Habib, N. et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020).
Nasrabady, S. E., Rizvi, B., Goldman, J. E. & Brickman, A. M. White matter changes in Alzheimer’s disease: a focus on myelin and oligodendrocytes. Acta Neuropathol. Commun. 6, 22 (2018).
Wu, Y. et al. Alterations of myelin morphology and oligodendrocyte development in early stage of Alzheimer’s disease mouse model. Neurosci. Lett. 642, 102–106 (2017).
Behrendt, G. et al. Dynamic changes in myelin aberrations and oligodendrocyte generation in chronic amyloidosis in mice and men. Glia 61, 273–286 (2013).
Keren-Shaul, H. et al. MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat. Protoc. 14, 1841–1862 (2019).
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Lake, B. B. et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2018).
Bost, P. et al. Host–viral infection maps reveal signatures of severe COVID-19 patients. Cell 181, 1475–1488 (2020).
Robinson, A. P., Rodgers, J. M., Goings, G. E. & Miller, S. D. Characterization of oligodendroglial populations in mouse demyelinating disease using flow cytometry: clues for MS pathogenesis. PLoS ONE 9, e107649 (2014).
Vicuña, L. et al. The serine protease inhibitor SerpinA3N attenuates neuropathic pain by inhibiting T cell-derived leukocyte elastase. Nat. Med. 21, 518–523 (2015).
Haile, Y. et al. Granzyme B-inhibitor serpina3n induces neuroprotection in vitro and in vivo. J. Neuroinflammation 12, 157 (2015).
Gadani, S. P., Walsh, J. T., Smirnov, I., Zheng, J. & Kipnis, J. The glia-derived alarmin IL-33 orchestrates the immune response and promotes recovery following CNS injury. Neuron 85, 703–709 (2015).
Bando, Y. et al. Kallikrein 6 secreted by oligodendrocytes regulates the progression of experimental autoimmune encephalomyelitis. Glia 66, 359–378 (2018).
Kwon, O.-C. et al. SGK1 inhibition in glia ameliorates pathologies and symptoms in Parkinson disease animal models. EMBO Mol. Med. 13, e13076 (2021).
Gupta, A. & Pulliam, L. Exosomes as mediators of neuroinflammation. J. Neuroinflammation 11, 68 (2014).
Pascual, M., Ibáñez, F. & Guerri, C. Exosomes as mediators of neuron–glia communication in neuroinflammation. Neural Regen. Res. 15, 796–801 (2020).
Janky, R. et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 10, e1003731 (2014).
Hashimoto, R. et al. Origins of oligodendrocytes in the cerebellum, whose development is controlled by the transcription factor, Sox9. Mech. Dev. 140, 25–40 (2016).
Stolt, C. C. et al. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689 (2003).
Lee, S.-H. et al. Trem2 restrains the enhancement of tau accumulation and neurodegeneration by β-amyloid pathology. Neuron 109, 1283–1301 (2021).
Stine, W. B., Jungbauer, L., Yu, C. & Ladu, M. J. Preparing synthetic Aβ in different aggregation states. Methods Mol. Biol. 670, 13–32 (2011).
Constantinescu, C. S., Farooqi, N., O’Brien, K. & Gran, B. Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS). Br. J. Pharm. 164, 1079 (2011).
Rangachari, M. & Kuchroo, V. K. Using EAE to better understand principles of immune function and autoimmune pathology. J. Autoimmun. 45, 31–39 (2013).
Falcão, A. M. et al. Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat. Med. 24, 1837–1844 (2018).
Wheeler, M. A. et al. MAFG-driven astrocytes promote CNS inflammation. Nature 578, 593–599 (2020).
Dulken, B. W. et al. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210 (2019).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Hasel, P., Rose, I. V. L., Sadick, J. S., Kim, R. D. & Liddelow, S. A. Neuroinflammatory astrocyte subtypes in the mouse brain. Nat. Neurosci. 24, 1475–1487 (2021).
Greenwald, N. F. et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565 (2022).
Illian, J., Penttinen, A., Stoyan, H. & Stoyan, D. Statistical Analysis and Modelling of Spatial Point Patterns. Statistical Analysis and Modelling of Spatial Point Patterns (Wiley, 2008).
Nagele, R. G. et al. Contribution of glial cells to the development of amyloid plaques in Alzheimer’s disease. Neurobiol. Aging 25, 663–674 (2004).
Chen, W. T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991 (2020).
Serrano-Pozo, A., Betensky, R. A., Frosch, M. P. & Hyman, B. T. Plaque-associated local toxicity increases over the clinical course of Alzheimer disease. Am. J. Pathol. 186, 375–384 (2016).
Yin, Z. et al. Immune hyperreactivity of Aβ plaque-associated microglia in Alzheimer’s disease. Neurobiol. Aging 55, 115–122 (2017).
Griciuc, A. et al. TREM2 acts downstream of CD33 in modulating microglial pathology in Alzheimer’s disease. Neuron 103, 820–835 (2019).
Rajala, T., Murrell, D. J. & Olhede, S. C. Detecting multivariate interactions in spatial point patterns with Gibbs models and variable selection. J. R. Stat. Soc. Ser. C. Appl. Stat. 67, 1237–1273 (2018).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Thrupp, N. et al. Single-nucleus RNA-seq is not suitable for detection of microglial activation genes in humans. Cell Rep. 32, 108189 (2020).
Chen, S. et al. Spatially resolved transcriptomics reveals unique gene signatures associated with human temporal cortical architecture and Alzheimer’s pathology. Preprint at https://www.biorxiv.org/content/10.1101/2021.07.07.451554v1 (2021).
Bin, J. M., Harris, S. N. & Kennedy, T. E. The oligodendrocyte-specific antibody ‘CC1’ binds Quaking 7. J. Neurochem. 139, 181–186 (2016).
Venegas, C. & Heneka, M. T. Danger-associated molecular patterns in Alzheimer’s disease. J. Leukoc. Biol. 101, 87–98 (2017).
Zhang, P. et al. Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci. 22, 719–728 (2019).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012).
Deczkowska, A. et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat. Commun. 8, 717 (2017).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Lai, L. et al. GSKB: a gene set database for pathway analysis in mouse. Preprint at https://www.biorxiv.org/content/10.1101/082511v1.full (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Acknowledgements
The research of I.A. is supported by the Seed Networks for the Human Cell Atlas of the Chan Zuckerberg Initiative, the Thompson Family Foundation Alzheimer’s Research Fund and the Adelis Foundation. I.A. is an Eden and Steven Romick Professorial Chair, supported by the HHMI International Scholar Award, a European Research Council Consolidator Grant (724471-HemTree2.0), an MRA Established Investigator Award (509044), the DFG (SFB/TRR167), the Ernest and Bonnie Beutler Research Program for Excellence in Genomic Medicine, the Helen and Martin Kimmel awards for innovative investigation and the SCA award of the Wolfson Foundation and Family Charitable Trust. Research in the M.S. laboratory is supported by Advanced European Research Council grants (741744); Israel Science Foundation (ISF) research grant 991/16; and ISF-Legacy Heritage Bio-Medical Science Partnership research grant 1354/15. We would like to thank the Adelis and Thompson Foundations for their generous support of our AD research. This work is also supported by awards K01-AG056673 and R56-066782–01 from the National Institute on Aging of the National Institutes of Health, R01-GM131399 (Q.M.) from the National Institute of General Medical Sciences, AARF-17–505009 (H.F.) from the Alzheimer’s Association and W81XWH1910309 (H.F.) from the US Department of Defense.
Author information
Authors and Affiliations
Contributions
M.S., I.A. and M.K. conceived the study. M.K and P.B. designed the experiments. M.K. conducted the animal work, isolated cells and generated single-cell sequencing libraries. M.K. and S.H. conducted the imaging experiments. M.K. and S.H. conducted the in vitro experiments, with assistance from R.H. P.B. conducted the data analysis for the sequencing, re-analysis of the published datasets and spatial analysis of the mouse LPS treatment Visium data. P.B. developed a new algorithmic approach for image analysis and analyzed imaging experiments. Y.C., S.C, Q.M. and H.F. conducted the Visium spatial transcriptomics on human brain samples and the data analysis. B.S. and B.B. provided the computational resources for analysis. M.K., M.S., P.B. and I.A. wrote the paper, with input from all the other authors.
Corresponding authors
Ethics declarations
Competing interests
M.S. serves as a consultant of Immunobrain Checkpoint Ltd.
Peer review
Peer review information
Nature Neuroscience thanks Sarah Jäkel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Quality control of CD45− libraries described in Fig. 1.
Extended data Fig. 1 associated to Fig. 1. (a) Distribution of total cellular unique molecular identifiers (UMI). Dashed line marks the threshold for analysis. (b) Distribution of total gene UMIs. Dashed line marks the threshold for analysis. (c) Distribution of the proportion of mitochondrial genes. Dashed line marks the threshold for analysis. (d) Comparison of the total cellular UMI distribution in cells from 5xFAD and WT mice (left panel) and the proportion of mitochondrial genes in cells from 5xAD and WT mice (right panel). P values were computed using a Kruskal-Wallis test. The box bounds the IQR. Line, median. Whiskers extend to a maximum of 1.5*IQR beyond the box. n = 18 independent mice (9 5xFAD, 9 WT) (e) Violin plot of known marker genes expression across the different cell clusters. (f) Comparison of the main cell-type proportions between 5xAD and WT mice.
Extended Data Fig. 2 Quality control of GalC+ libraries described in Fig. 2.
Extended data Fig. 2 associated to Fig. 2. (a) Gating strategy used to enrich for oligodendrocytes. (b) Proportion of oligodendrocytes isolated in each sequenced plate. Large bars correspond to the median and small bars to IQR. n = 48 384-well plates (c) Number of oligodendrocytes sequenced for each 5xAD and WT mice (left panel) and across ages (right panel). Large bars correspond to the median and small bars to IQR. n = 33 independent mice (17 5xFAD, 16 WT) (d) Proportion of cluster 14 in WT and 5xFAD mice across ages. Large bars correspond to the median and small bars to IQR. n = 33 independent mice (across ages: 6–8 m; n = 4 5xFAD, 4 WT, 10–11 m; n = 6 5xFAD, 5 WT, 15 m; n = 3 5xFAD, 3 WT, 24 m; n = 4 5xFAD, 4 WT) (e) Volcano plot corresponding to the differential expression analysis between DOL-like and the rest of oligodendrocytes as identified by Zhou et al.11. DOL genes are colored in orange. (f) Proportion of DOL-like among oligodendrocytes between 5xFAD and WT mice in the data by Zhou et al.11. P-value was computed using the Kruskal-Wallis test.
Extended Data Fig. 3 Cell type annotation of dataset described in Fig. 3 and culture quality control.
Extended data Fig. 3 associated to Fig. 3. (a) Spearman correlation between the mean transcriptomic profiles of the cell clusters identified in the dataset from Lee et al. (b) Violin plot of known marker genes across the different cell clusters. (c) Violin plot of known DAM marker gene expression across the different microglia clusters after refined clustering. (d) Representative bright-field microscopy image of the primary oligodendrocyte culture; scale bar corresponds to 50 μm. Representative results from 12 independent experiments.
Extended Data Fig. 4 GSEA and quality control of datasets described in Fig. 4.
Extended data Fig. 4 associated to Fig. 4. (a) GSEA analysis plot corresponding to the acute EAE dataset. (b) GSEA analysis plot corresponding to the multiphasic EAE dataset. (c) GSEA analysis plot corresponding to the aging SVZ dataset. (d) score of topic number 8, corresponding to DOL-like signature, in LPS-stimulated and control samples. n = 6 independent samples (3 LPS-stimulated, 3 control). Thick line corresponds to the median, the bottom and upper limits of the box to the first and third quartile, respectively. The lower and upper whiskers correspond to the lowest and highest values respectively within the range of the first (third) quartile minus (plus) 1.5 times the Interquartile range. (e) GSEA analysis plot of the DOL signature in topic 8. (f) Serpina3n expression (transcripts per thousand) in LPS-treated and control mice. n = 6 independent samples (3 LPS-stimulated, 3 control). p-value was computed by performing a Gene Set Enrichment Analysis as described by Subramanian et al56. Thick line corresponds to the median, the bottom and upper limits of the box to the first and third quartile, respectively. The lower and upper whiskers correspond to the lowest and highest values respectively within the range of the first (third) quartile minus (plus) 1.5 times the Interquartile range. (g) Intensity of the oligodendrocyte signature across the sections of the three LPS treated mice.
Extended Data Fig. 5 Quality control of image analysis described in Fig. 5.
Extended data Fig. 5 associated to Fig. 5. (a) Separated channels corresponding to the 5xFAD brain sample in Fig. 5a; scale bar corresponds to 20 μm. (b) Distribution of cell size across mice samples. The box bounds the IQR. Line, median. Whiskers extend to a maximum of 1.5*IQR beyond the box. n = 6 independent samples (4 5xFAD, 2 WT). (c) Distribution of OLIG2 intensity across mouse samples. The box bounds the IQR. Line, median. Whiskers extend to a maximum of 1.5*IQR beyond the box. n = 6 independent samples (4 5xFAD, 2 WT). (d) Distribution of OLIG2 intensity and estimated threshold (vertical red line). (e) Proportion of OLIG2+ cells across mouse samples. (f) Number of plaques in 5xFAD (n = 4) and WT (n = 2) mice. (g) Distribution of SERPINA3N intensity and the estimated threshold (vertical red line). (h) The 15 most contributing genes to the macrophage signature. (i) The 15 most contributing genes to the inflammatory signature. (j) Separated channels corresponding to the postmortem AD brain sample in Fig. 5i; scale bar corresponds to 50 μm. (k) Distribution of cell size across human samples. The box bounds the IQR. Line, median. Whiskers extend to a maximum of 1.5*IQR beyond the box. n = 16 independent samples (8 AD, 8 NDC). (l) Distribution of SERPINA3 intensity and the estimated threshold (vertical red line) for the two different batches of samples.
Supplementary information
Supplementary Table 1
Details of mice used in the study
Supplementary table 2
DEGs in 5xFAD compared to WT, according to different cell types. P values were computed using a binomial regression with complementary log–log link function (Methods)
Supplementary table 3
Details of postmortem AD patients brain samples used in the study
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kenigsbuch, M., Bost, P., Halevi, S. et al. A shared disease-associated oligodendrocyte signature among multiple CNS pathologies. Nat Neurosci 25, 876–886 (2022). https://doi.org/10.1038/s41593-022-01104-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01104-7
This article is cited by
-
Deletion of Slc9a1 in Cx3cr1+ cells stimulated microglial subcluster CREB1 signaling and microglia-oligodendrocyte crosstalk
Journal of Neuroinflammation (2024)
-
SRT-Server: powering the analysis of spatial transcriptomic data
Genome Medicine (2024)
-
Immunological aspects of central neurodegeneration
Cell Discovery (2024)
-
Oligodendrocyte–axon metabolic coupling is mediated by extracellular K+ and maintains axonal health
Nature Neuroscience (2024)
-
Cell type-specific roles of APOE4 in Alzheimer disease
Nature Reviews Neuroscience (2024)