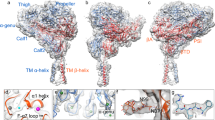
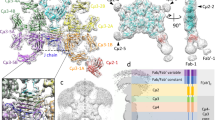
Abstract
Integrins are conformationally flexible cell surface receptors that survey the extracellular environment for their cognate ligands. Interactions with ligands are thought to be linked to global structural rearrangements involving transitions between bent, extended-closed and extended-open forms. Thus far, structural details are lacking for integrins in the extended conformations due to extensive flexibility between the headpiece and legs in this conformation. Here we present single-particle electron cryomicroscopy structures of human αvβ8 integrin in the extended-closed conformation, which has been considered to be a low-affinity intermediate. Our structures show the headpiece rotating about a flexible αv knee, suggesting a ligand surveillance mechanism for integrins in their extended-closed form. Our model predicts that the extended conformation is mainly stabilized by an interface formed between flexible loops in the upper and lower domains of the αv leg. Confirming these findings with the αvβ3 integrin suggests that our model of stabilizing the extended-closed conformation is generalizable to other integrins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hynes, R. O. The emergence of integrins: a personal and historical perspective. Matrix Biol. 23, 333–340 (2004).
Campbell, I. D. & Humphries, M. J. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 3, a004994 (2011).
Takagi, J., Petre, B. M., Walz, T. & Springer, T. A. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110, 599–611 (2002).
Xiong, J. P. et al. Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment. J. Cell Biol. 186, 589–600 (2009).
Luo, B. H., Carman, C. V. & Springer, T. A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 (2007).
Minagawa, S. et al. Selective targeting of TGF-β activation to treat fibroinflammatory airway disease. Sci. Transl. Med. 6, 241ra79 (2014).
Wang, J. et al. Atypical interactions of integrin αVβ8 with pro-TGF-β1. Proc. Natl. Acad. Sci. USA. 114, E4168–E4174 (2017).
Cheng, Y. Single-particle cryo-EM at crystallographic resolution. Cell 161, 450–457 (2015).
Cheng, Y., Grigorieff, N., Penczek, P. A. & Walz, T. A primer to single-particle cryo-electron microscopy. Cell 161, 438–449 (2015).
Wu, S. et al. Fabs enable single particle cryoEM studies of small proteins. Structure 20, 582–592 (2012).
Adams, P. D. et al. a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Lyskov, S. et al. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS One 8, e63906 (2013).
Xiong, J. P. et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155 (2002).
Mu, D. et al. The integrin αvβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J. Cell Biol. 157, 493–507 (2002).
Ozawa, A. et al. Molecular basis of the ligand binding specificity of αvβ8 integrin. J. Biol. Chem. 291, 11551–11565 (2016).
Robertson, I. B. et al. Latent TGF-β-binding proteins. Matrix Biol. 47, 44–53 (2015).
Tran, D. Q. et al. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proc. Natl. Acad. Sci. USA. 106, 13445–13450 (2009).
Nishimura, S. L. Integrin-mediated transforming growth factor-beta activation, a potential therapeutic target in fibrogenic disorders. Am. J. Pathol. 175, 1362–1370 (2009).
Kamata, T. et al. Critical cysteine residues for regulation of integrin alphaIIbbeta3 are clustered in the epidermal growth factor domains of the beta3 subunit. Biochem. J. 378, 1079–1082 (2004).
Mor-Cohen, R. et al. Disulfide bond disruption by a beta 3-Cys549Arg mutation in six Jordanian families with Glanzmann thrombasthenia causes diminished production of constitutively active alpha IIb beta 3. Thromb. Haemost. 98, 1257–1265 (2007).
Smagghe, B. J., Huang, P. S., Ban, Y. E., Baker, D. & Springer, T. A. Modulation of integrin activation by an entropic spring in the β-knee. J. Biol. Chem. 285, 32954–32966 (2010).
Frelinger, A. L. III, Du, X. P., Plow, E. F. & Ginsberg, M. H. Monoclonal antibodies to ligand-occupied conformers of integrin alpha IIb beta 3 (glycoprotein IIb-IIIa) alter receptor affinity, specificity, and function. J. Biol. Chem. 266, 17106–17111 (1991).
Honda, S. et al. Topography of ligand-induced binding sites, including a novel cation-sensitive epitope (AP5) at the amino terminus, of the human integrin beta 3 subunit. J. Biol. Chem. 270, 11947–11954 (1995).
Stanley, P. Chinese hamster ovary cell mutants with multiple glycosylation defects for production of glycoproteins with minimal carbohydrate heterogeneity. Mol. Cell. Biol. 9, 377–383 (1989).
Nishimura, S. L., Sheppard, D. & Pytela, R. Integrin αvβ8: interaction with vitronectin and functional divergence of the beta 8 cytoplasmic domain. J. Biol. Chem. 269, 28708–28715 (1994).
Shi, M. et al. Latent TGF-β structure and activation. Nature 474, 343–349 (2011).
Booth, D.S., Avila-Sakar, A. & Cheng, Y. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J. Vis. Exp. 3227 https://doi.org/10.3791/3227 (2011).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Li, X., Zheng, S., Agard, D. A. & Cheng, Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 192, 174–178 (2015).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Brubaker, M. A. & Fleet, D. J. Building proteins in a day: efficient 3D molecular structure estimation with electron cryomicroscopy. IEEE Trans. Pattern Anal. Mach. Intell. 39, 706–718 (2017).
Grigorieff, N. Frealign: an exploratory tool for single-particle Cryo-EM. Methods Enzymol. 579, 191–226 (2016).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
DeLano, W. L. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92 (2002).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics47, 5.6.1–5.6.32 https://doi.org/10.1002/0471250953.bi0506s47 (2014).
Singh, A. et al. Extension and validation of the GLYCAM force field parameters for modeling glycosaminoglycans. Can. J. Chem. 94, 927–935 (2016).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Weinacker, A. et al. Role of the integrin αvβ6 in cell attachment to fibronectin. Heterologous expression of intact and secreted forms of the receptor. J. Biol. Chem. 269, 6940–6948 (1994).
Gline, S. E., Cambier, S., Govaerts, C. & Nishimura, S. L. A. A 50-Å separation of the integrin alpha v beta 3 extracellular domain C termini reveals an intermediate activation state. J. Biol. Chem. 279, 54567–54572 (2004).
Notredame, C., Higgins, D. G. & Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000).
Acknowledgements
We thank M. Braunfeld for supporting the cryo-EM facility at UCSF. LIBS antibodies were a gift from M. Ginsberg (University of California San Diego). This work was supported in part by grants the NIH (U54HL119893 and R01HL113032 to S.L.N.; R01HL134183 to S.L.N. and Y.C.; R01GM098672, S10OD020054 and S10OD021741 to Y.C.; and P41CA196276 to J.M.) and from the University of California Office of the President Tobacco-Related Disease Research Program to S.L.N. Y.C. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
A.C., M.G.C. and S.W. performed cryo-EM and structural biology. A.C. and M.G.C. performed ns-EM. A.C. and S.I. performed biochemical experiments. S.I., A.C. and S.L.N. designed, generated and characterized mutant integrins. A.C., M.G.C., S.I., S.W., S.L.N. and Y.C. conceived experiments and wrote the manuscript. J.M., J.L., J.B. and S.L.N. produced, characterized, cloned and engineered monoclonal antibodies.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Models of integrin conformational changes and activation mechanisms.
a, The individual leg, head and sub-domains are shown using common nomenclature. b, In the switchblade model, the bent conformation moves through an extended-closed conformation to an extended-open conformation. In this model, the bent conformation is low-affinity, the extend-closed is a low-affinity intermediate, and the extended-open is the high-affinity fully active conformation. c, In the bent model, affinity regulation occurs solely in the bent conformation. d, In the case of αvβ8, affinity regulation must occur in the extended-closed conformation. In all models, affinity changes corresponding to movements in the headpiece are indicated by pink arrows.
Supplementary Figure 2 Cryo-EM micrograph, local resolution estimates and orientation distribution of the αvβ8-8B8-68 complex.
a, Representative electron micrograph and b, 2D class averages of frozen hydrated αvβ8-8B8-68 complex. The scale bar is 500 Å in the micrograph, and 200 Å in 2D class averages. c, Fourier Shell Correlation (FSC) curves of whole αvβ8-8B8-68 complex (blue solid line) and headpiece alone (blue dashed line). d, FSC curve between the density map and the fitted atomic model of αvβ8-8B8-68 complex. e, Local resolution of αvβ8-8B8-68 complex (left) and angular distributions of all particles used for calculating 3D reconstruction (right). The figures are color coded as indicated. f, Local resolution of αvβ8-8B8-68 complex with focused alignment in its headpiece (left) and angular distributions of all particles used for calculating 3D reconstruction (right), which is almost identical to the angular distribution of whole molecule reconstruction.
Supplementary Figure 3 Processing schematic for αvβ8-8B8-68 complex.
A schematic flowchart showing the classification scheme of αvβ8-8B8-68 complex. Particle numbers at each step and for each class are indicated. 3D reconstructions of six out of eight subclasses reached sub-nanometer resolution, despite small particle numbers for each class. Local resolution for each subclass are color coded as indicated. Two classes that did not yield comparable reconstructions were excluded from further analysis.
Supplementary Figure 4 Contacts between the αv thigh and calf-1 domains stabilize the extended conformation.
a and b, Same views of density maps for all subclasses show the progressive loss of contact between αv-leg and β8 upper leg (a), and disappearance of the β8 lower leg (b). The color code is: αv-green; β8-blue.
Supplementary Figure 5 Extension of αvβ3 does not change its intrinsic ligand affinity; integrin β leg sequence alignment.
a, Receptor binding of αvβ3 ectodomains (WT and αvc-cβ3) to vitronectin-N in basal (Ca2+) or activating (Mn2+) cation conditions (n = 3, ± s.e.m.). b, Receptor binding of αvβ8 ectodomains (WT and αvc-cβ8) to L-TGF-β (n = 3, ± s.e.m.). c, Sequence alignment for the leg of all β integrins pairing with αv. Yellow highlights conserved cysteines in the β legs, notable exceptions in the β8 leg are highlighted in blue.
Supplementary information
Supplementary Video 1
Molecular animation of the headpiece movement. Animated movie of ribbon models of the six αvβ8-8B8-68 subclasses aligned to the αv-calf-1,2 region of subclass (iv). The movie depicts αvβ8-8B8-68 in the membrane and moves sequentially through subclasses (i), (ii), (iii), (v) and (vi), each time moving back through subclass (iv).
Rights and permissions
About this article
Cite this article
Cormier, A., Campbell, M.G., Ito, S. et al. Cryo-EM structure of the αvβ8 integrin reveals a mechanism for stabilizing integrin extension. Nat Struct Mol Biol 25, 698–704 (2018). https://doi.org/10.1038/s41594-018-0093-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0093-x
This article is cited by
-
Organization, dynamics and mechanoregulation of integrin-mediated cell–ECM adhesions
Nature Reviews Molecular Cell Biology (2023)
-
Molecular mechanisms of catch bonds and their implications for platelet hemostasis
Biophysical Reviews (2023)
-
Specificity of TGF-β1 signal designated by LRRC33 and integrin αVβ8
Nature Communications (2022)
-
Suppression of the fibrotic encapsulation of silicone implants by inhibiting the mechanical activation of pro-fibrotic TGF-β
Nature Biomedical Engineering (2021)
-
General structural features that regulate integrin affinity revealed by atypical αVβ8
Nature Communications (2019)