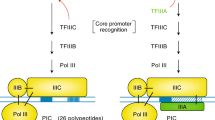
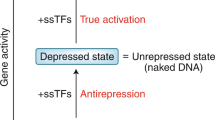
Abstract
The discovery of the three eukaryotic nuclear RNA polymerases paved the way for serious biochemical investigations of eukaryotic transcription and the identification of eukaryotic transcription factors. Here we describe this adventure from our vantage point, with a focus on the hunt for factors that regulate elongation by RNA polymerase II.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Thummel, C. S., Burtis, K. C. & Hogness, D. S. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell 61, 101–111 (1990).
Jonkers, I., Kwak, H. & Lis, J. T. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3, e02407 (2014).
Saponaro, M. et al. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell 157, 1037–1049 (2014).
Saunders, A., Core, L. J. & Lis, J. T. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7, 557–567 (2006).
Tamm, I., Kikuchi, T., Darnell, J. E. Jr. & Salditt-Georgieff, M. Short capped hnRNA precursor chains in HeLa cells: continued synthesis in the presence of 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole. Biochemistry 19, 2743–2748 (1980).
Jonkers, I. & Lis, J. T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 (2015).
Sekimizu, K., Kobayashi, N., Mizuno, D. & Natori, S. Purification of a factor from Ehrlich ascites tumor cells specifically stimulating RNA polymerase II. Biochemistry 15, 5064–5070 (1976).
Fish, R. N. & Kane, C. M. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577, 287–307 (2002).
Reines, D., Ghanouni, P., Li, Q. Q. & Mote, J. Jr. The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J. Biol. Chem. 267, 15516–15522 (1992).
Reines, D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J. Biol. Chem. 267, 3795–3800 (1992).
Izban, M. G. & Luse, D. S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′→5′ direction in the presence of elongation factor SII. Genes Dev. 6, 1342–1356 (1992).
Churchman, L. S. & Weissman, J. S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 (2011).
Sheridan, R. M., Fong, N., D’Alessandro, A. & Bentley, D. L. widespread backtracking by RNA Pol II is a major effector of gene activation, 5′ pause release, termination, and transcription elongation rate. Mol. Cell 73, 107–118.e4 (2019).
Lemay, J. F. et al. The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat. Struct. Mol. Biol. 21, 919–926 (2014).
Nechaev, S. et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335–338 (2010).
Adelman, K. et al. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17, 103–112 (2005).
Sigurdsson, S., Dirac-Svejstrup, A. B. & Svejstrup, J. Q. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol. Cell 38, 202–210 (2010).
Gamba, P. & Zenkin, N. Transcription fidelity and its roles in the cell. Curr. Opin. Microbiol. 42, 13–18 (2018).
Irvin, J. D. et al. A genetic assay for transcription errors reveals multilayer control of RNA polymerase II fidelity. PLoS Genet. 10, e1004532 (2014).
Gout, J. F. et al. The landscape of transcription errors in eukaryotic cells. Sci. Adv. 3, e1701484 (2017).
Sawadogo, M. & Sentenac, A. RNA polymerase B (II) and general transcription factors. Annu. Rev. Biochem. 59, 711–754 (1990).
Conaway, R. C., Garrett, K. P., Hanley, J. P. & Conaway, J. W. Mechanism of promoter selection by RNA polymerase II: mammalian transcription factors α and β γ promote entry of polymerase into the preinitiation complex. Proc. Natl Acad. Sci. USA 88, 6205–6209 (1991).
Bradsher, J. N., Jackson, K. W., Conaway, R. C. & Conaway, J. W. RNA polymerase II transcription factor SIII. I. Identification, purification, and properties. J. Biol. Chem. 268, 25587–25593 (1993).
Bradsher, J. N., Tan, S., McLaury, H.-J., Conaway, J. W. & Conaway, R. C. RNA polymerase II transcription factor SIII. II. Functional properties and role in RNA chain elongation. J. Biol. Chem. 268, 25594–25603 (1993).
Shilatifard, A., Lane, W. S., Jackson, K. W., Conaway, R. C. & Conaway, J. W. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271, 1873–1876 (1996).
Simone, F. et al. EAF1, a novel ELL-associated factor that is delocalized by expression of the MLL-ELL fusion protein. Blood 98, 201–209 (2001).
Simone, F., Luo, R. T., Polak, P. E., Kaberlein, J. J. & Thirman, M. J. ELL-associated factor 2 (EAF2), a functional homolog of EAF1 with alternative ELL binding properties. Blood 101, 2355–2362 (2003).
Kong, S. E., Banks, C. A. S., Shilatifard, A., Conaway, J. W. & Conaway, R. C. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc. Natl Acad. Sci. USA 102, 10094–10098 (2005).
Price, D. H., Sluder, A. E. & Greenleaf, A. L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol. 9, 1465–1475 (1989).
Bengal, E., Flores, O., Krauskopf, A., Reinberg, D. & Aloni, Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol. Cell. Biol. 11, 1195–1206 (1991).
Shilatifard, A., Conaway, R. C. & Conaway, J. W. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72, 693–715 (2003).
Yan, Q., Moreland, R. J., Conaway, J. W. & Conaway, R. C. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J. Biol. Chem. 274, 35668–35675 (1999).
Joo, Y. J., Ficarro, S. B., Chun, Y., Marto, J. A. & Buratowski, S. In vitro analysis of RNA polymerase II elongation complex dynamics. Genes Dev. 33, 578–589 (2019).
Gerber, M. et al. In vivo requirement of the RNA polymerase II elongation factor elongin A for proper gene expression and development. Mol. Cell. Biol. 24, 9911–9919 (2004).
Eissenberg, J. C. et al. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc. Natl Acad. Sci. USA 99, 9894–9899 (2002).
Mitani, K. et al. Nonredundant roles of the elongation factor MEN in postimplantation development. Biochem. Biophys. Res. Commun. 279, 563–567 (2000).
Yasukawa, T. et al. Transcriptional elongation factor elongin A regulates retinoic acid-induced gene expression during neuronal differentiation. Cell Rep. 2, 1129–1136 (2012).
Gerber, M. et al. Regulation of heat shock gene expression by RNA polymerase II elongation factor, Elongin A. J. Biol. Chem. 280, 4017–4020 (2005).
Kawauchi, J. et al. Transcriptional properties of mammalian elongin A and its role in stress response. J. Biol. Chem. 288, 24302–24315 (2013).
Chopra, V. S., Hong, J. W. & Levine, M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr. Biol. 19, 688–693 (2009).
Ardehali, M. B. et al. Polycomb repressive complex 2 methylates Elongin A to regulate transcription. Mol. Cell 68, 872–884.e6 (2017).
Kamura, T. et al. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276, 29748–29753 (2001).
Selth, L. A., Sigurdsson, S. & Svejstrup, J. Q. Transcript elongation by RNA polymerase II. Annu. Rev. Biochem. 79, 271–293 (2010).
Kamura, T. et al. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12, 3872–3881 (1998).
Kamura, T. et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657–661 (1999).
Okumura, F., Matsuzaki, M., Nakatsukasa, K. & Kamura, T. The role of Elongin BC-containing ubiquitin ligases. Front. Oncol. 2, 10 (2012).
Ribar, B., Prakash, L. & Prakash, S. ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells. Mol. Cell. Biol. 27, 3211–3216 (2007).
Yasukawa, T. et al. Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 27, 3256–3266 (2008).
Harreman, M. et al. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc. Natl Acad. Sci. USA 106, 20705–20710 (2009).
Weems, J. C. et al. Assembly of the Elongin A ubiquitin ligase is regulated by genotoxic and other stresses. J. Biol. Chem. 290, 15030–15041 (2015).
Weems, J. C. et al. Cockayne syndrome B protein regulates recruitment of the Elongin A ubiquitin ligase to sites of DNA damage. J. Biol. Chem. 292, 6431–6437 (2017).
Boetefuer, E. L., Lake, R. J. & Fan, H. Y. Mechanistic insights into the regulation of transcription and transcription-coupled DNA repair by Cockayne syndrome protein B. Nucleic Acids Res. 46, 7471–7479 (2018).
Wang, W., Xu, J., Chong, J. & Wang, D. Structural basis of DNA lesion recognition for eukaryotic transcription-coupled nucleotide excision repair. DNA Repair (Amst.) 71, 43–55 (2018).
Dahmus, M. E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271, 19009–19012 (1996).
Marshall, N. F. & Price, D. H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270, 12335–12338 (1995).
Zhu, Y. et al. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11, 2622–2632 (1997).
Peng, J., Marshall, N. F. & Price, D. H. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273, 13855–13860 (1998).
Wada, T. et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 (1998).
Yamaguchi, Y. et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 (1999).
Swanson, M. S. & Winston, F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132, 325–336 (1992).
Fujinaga, K. et al. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24, 787–795 (2004).
Yamada, T. et al. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21, 227–237 (2006).
Lin, C. et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 37, 429–437 (2010).
He, N. et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 38, 428–438 (2010).
Sobhian, B. et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell 38, 439–451 (2010).
Takahashi, H. et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 (2011).
Lin, C. et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 25, 1486–1498 (2011).
Galbraith, M. D. et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 153, 1327–1339 (2013).
Wan, L. et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269 (2017).
Liang, K. et al. Targeting processive transcription elongation via SEC disruption for MYC-induced cancer therapy. Cell 175, 766–779.e17 (2018).
Marschalek, R. Mixed lineage leukemia: roles in human malignancies and potential therapy. FEBS J. 277, 1822–1831 (2010).
Yokoyama, A., Lin, M., Naresh, A., Kitabayashi, I. & Cleary, M. L. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17, 198–212 (2010).
Lin, C., Garruss, A. S., Luo, Z., Guo, F. & Shilatifard, A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell 152, 144–156 (2013).
Peterlin, B. M. & Price, D. H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006).
Lens, Z. et al. Solution structure of the N-terminal domain of Mediator subunit MED26 and molecular characterization of its interaction with EAF1 and TAF7. J. Mol. Biol. 429, 3043–3055 (2017).
He, N. et al. Human polymerase-associated factor complex (PAFc) connects the super elongation complex (SEC) to RNA polymerase II on chromatin. Proc. Natl Acad. Sci. USA 108, E636–E645 (2011).
Yadav, D., Ghosh, K., Basu, S., Roeder, R. G. & Biswas, D. Multivalent role of human TFIID in recruiting elongation components at the promoter-proximal region for transcriptional control. Cell Rep. 26, 1303–1317.e7 (2019).
Li, Y. et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell 62, 181–193 (2016).
Smith, E. R. et al. The little elongation complex regulates small nuclear RNA transcription. Mol. Cell 44, 954–965 (2011).
Hu, D. et al. The little elongation complex functions at initiation and elongation phases of snRNA gene transcription. Mol. Cell 51, 493–505 (2013).
Egloff, S. et al. The 7SK snRNP associates with the little elongation complex to promote snRNA gene expression. EMBO J. 36, 934–948 (2017).
Takahashi, H. et al. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat. Commun. 6, 5941 (2015).
Acknowledgements
We thank J. Weems for help with figures and all of the many colleagues who have contributed to our laboratory’s research on Pol II elongation over the years. Because of length limitations, we have not provided exhaustive references and in some cases have cited reviews rather than primary papers. We apologize to those whose contributions to the primary literature were not cited.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Conaway, R.C., Conaway, J.W. The hunt for RNA polymerase II elongation factors: a historical perspective. Nat Struct Mol Biol 26, 771–776 (2019). https://doi.org/10.1038/s41594-019-0283-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0283-1
This article is cited by
-
Causes and consequences of RNA polymerase II stalling during transcript elongation
Nature Reviews Molecular Cell Biology (2021)