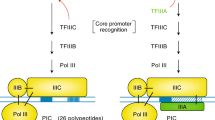
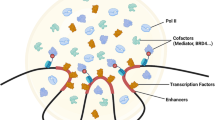
Abstract
Transcription regulation is critical to organism development and homeostasis. Control of expression of the 20,000 genes in human cells requires many hundreds of proteins acting through sophisticated multistep mechanisms. In this Historical Perspective, I highlight the progress that has been made in elucidating eukaryotic transcriptional mechanisms through an array of disciplines and approaches, and how this concerted effort has been driven by the development of new technologies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Roeder, R. G. & Rutter, W. J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature 224, 234–237 (1969).
Roeder, R.G. 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-019-0287-x (2019).
Sawadogo, M. & Roeder, R. G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc. Natl. Acad. Sci. USA 82, 4394–4398 (1985).
Matsui, T., Segall, J., Weil, P. A. & Roeder, R. G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J. Biol. Chem. 255, 11992–11996 (1980).
Burley, S. K. & Roeder, R. G. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769–799 (1996).
Kornberg, A. Ten commandments: lessons from the enzymology of DNA replication. J. Bacteriol. 182, 3613–3618 (2000).
Ruddle, F. H. A new era in mammalian gene mapping: somatic cell genetics and recombinant DNA methodologies. Nature 294, 115–120 (1981).
Weeks, J. R., Coulter, D. E. & Greenleaf, A. L. Immunological studies of RNA polymerase II using antibodies to subunits of Drosophila and wheat germ enzyme. J. Biol. Chem. 257, 5884–5892 (1982).
Gilmour, D. S. & Lis, J. T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 6, 3984–3989 (1986).
Rougvie, A. E. & Lis, J. T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54, 795–804 (1988).
Rasmussen, E. B. & Lis, J. T. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. USA 90, 7923–7927 (1993).
Kao, S. Y., Calman, A. F., Luciw, P. A. & Peterlin, B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330, 489–493 (1987).
Eick, D., Kohlhuber, F., Wolf, D. A. & Strobl, L. J. Activation of pausing RNA polymerases by nuclear run-on experiments. Anal. Biochem. 218, 347–351 (1994).
Krumm, A., Meulia, T., Brunvand, M. & Groudine, M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 6, 2201–2213 (1992).
Chodosh, L. A., Fire, A., Samuels, M. & Sharp, P. A. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 264, 2250–2257 (1989).
Yamaguchi, Y., Wada, T. & Handa, H. Interplay between positive and negative elongation factors: drawing a new view of DRB. Genes Cells 3, 9–15 (1998).
Marshall, N. F. & Price, D. H. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell. Biol. 12, 2078–2090 (1992).
Wada, T. et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 (1998).
Yamaguchi, Y. et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 (1999).
Wu, C. H. et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17, 1402–1414 (2003).
Li, J. et al. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol. Cell 50, 711–722 (2013).
Vos, S. M. et al. Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560, 607–612 (2018).
Winston, F., Chaleff, D. T., Valent, B. & Fink, G. R. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107, 179–197 (1984).
Muse, G. W. et al. RNA polymerase is poised for activation across the genome. Nat. Genet. 39, 1507–1511 (2007).
Zeitlinger, J. et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 (2007).
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008).
Core, L. J. et al. Defining the status of RNA polymerase at promoters. Cell Rep. 2, 1025–1035 (2012).
Jonkers, I., Kwak, H. & Lis, J. T. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3, e02407 (2014).
Mayran, A. & Drouin, J. Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 293, 13795–13804 (2018).
Pugh, B. F. & Venters, B. J. Genomic Organization of Human Transcription Initiation Complexes. PLoS One 11, e0149339 (2016).
Kouzine, F. et al. Global regulation of promoter melting in naive lymphocytes. Cell 153, 988–999 (2013).
Peterlin, B. M. & Price, D. H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006).
Gressel, S. et al. CDK9-dependent RNA polymerase II pausing controls transcription initiation. eLife 6, e29736 (2017).
Shao, W. & Zeitlinger, J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 49, 1045–1051 (2017).
Boettiger, A. N. & Levine, M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325, 471–473 (2009).
Duarte, F. M. et al. Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev. 30, 1731–1746 (2016).
Vihervaara, A. et al. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat. Commun. 8, 255 (2017).
Ptashne, M. & Gann, A. Transcriptional activation by recruitment. Nature 386, 569–577 (1997).
Kwak, H., Fuda, N. J., Core, L. J. & Lis, J. T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013).
Churchman, L. S. & Weissman, J. S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 (2011).
Wissink, E.M., Vihervaara, A., Tippens, N.D. & Lis, J.T. Nascent RNA analyses: tracking transcription and its regulation. Nat. Rev. Genet. https://doi.org/10.1038/s41576-019-0159-6 (2019).
Kruesi, W. S., Core, L. J., Waters, C. T., Lis, J. T. & Meyer, B. J. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2, e00808 (2013).
Nechaev, S. et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327, 335–338 (2010).
Schwalb, B. et al. TT-seq maps the human transient transcriptome. Science 352, 1225–1228 (2016).
Rhee, H. S. & Pugh, B. F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017).
Cui, K. & Zhao, K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol. Biol. 833, 413–419 (2012).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Kouzine, F. et al. Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome. Cell Syst. 4, 344–356 e7 (2017).
Mailler, E., Paillart, J. C., Marquet, R., Smyth, R. P. & Vivet-Boudou, V. The evolution of RNA structural probing methods: From gels to next-generation sequencing. Wiley Interdiscip. Rev. RNA 10, e1518 (2019).
Hannon, G. J. & Rossi, J. J. Unlocking the potential of the human genome with RNA interference. Nature 431, 371–378 (2004).
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Natsume, T. & Kanemaki, M. T. Conditional Degrons for Controlling Protein Expression at the Protein Level. Annu. Rev. Genet. 51, 83–102 (2017).
McDaniel, S. L. et al. Continued Activity of the Pioneer Factor Zelda Is Required to Drive Zygotic Genome Activation. Mol. Cell 74, 185–195.e4 (2019).
Bensaude, O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2, 103–108 (2011).
Shi, H., Hoffman, B. E. & Lis, J. T. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl. Acad. Sci. USA 96, 10033–10038 (1999).
Blau, J. et al. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16, 2044–2055 (1996).
Mahat, D. B., Salamanca, H. H., Duarte, F. M., Danko, C. G. & Lis, J. T. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Mol. Cell 62, 63–78 (2016).
Schoenfelder, S. & Fraser, P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Banerji, J., Rusconi, S. & Schaffner, W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981).
Gasperini, M. et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 176, 377–390.e19 (2019).
Fulco, C. P. et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016).
Cramer, P., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876 (2001).
Kostrewa, D. et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009).
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Patel, A. B. et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872 (2018).
Vos, S. M., Farnung, L., Urlaub, H. & Cramer, P. Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606 (2018).
Laird, C. D. & Chooi, W. Y. Morphology of transcription units in Drosophila melanogaster. Chromosoma 58, 193–218 (1976).
Zobeck, K. L., Buckley, M. S., Zipfel, W. R. & Lis, J. T. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol. Cell 40, 965–975 (2010).
Mir, M. et al. Single Molecule Imaging in Live Embryos Using Lattice Light-Sheet Microscopy. Methods Mol. Biol. 1814, 541–559 (2018).
Conic, S. et al. Imaging of native transcription factors and histone phosphorylation at high resolution in live cells. J. Cell Biol. 217, 1537–1552 (2018).
Kim, T. K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Core, L. J. et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320 (2014).
Henriques, T. et al. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 32, 26–41 (2018).
Arnold, C. D. et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (2013).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018).
Chen, H. et al. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 50, 1296–1303 (2018).
Roeder, R. G. Multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in Xenopus laevis. Isolation and partial characterization. J. Biol. Chem. 249, 241–248 (1974).
Zawel, L. & Reinberg, D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog. Nucleic Acid Res. Mol. Biol. 44, 67–108 (1993).
Peterson, C. L. & Tamkun, J. W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem. Sci. 20, 143–146 (1995).
Grunstein, M. & Hogness, D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl. Acad. Sci. USA 72, 3961–3965 (1975).
Kadonaga, J. T., Carner, K. R., Masiarz, F. R. & Tjian, R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51, 1079–1090 (1987).
Lander, E. S. Initial impact of the sequencing of the human genome. Nature 470, 187–197 (2011).
Fuda, N. J., Ardehali, M. B. & Lis, J. T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461, 186–192 (2009).
Acknowledgements
The author thanks members of his lab for valuable comments. The work is supported by US Public Health Service Award R01 GM025232 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Peer review information: Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lis, J.T. A 50 year history of technologies that drove discovery in eukaryotic transcription regulation. Nat Struct Mol Biol 26, 777–782 (2019). https://doi.org/10.1038/s41594-019-0288-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0288-9
This article is cited by
-
Cross-regulome profiling of RNA polymerases highlights the regulatory role of polymerase III on mRNA transcription by maintaining local chromatin architecture
Genome Biology (2022)
-
Altered macronutrient composition and genetics influence the complex transcriptional network associated with adiposity in the Collaborative Cross
Genes & Nutrition (2022)
-
Shape of promoter antisense RNAs regulates ligand-induced transcription activation
Nature (2021)