Featured
-
-
Article
| Open Access Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site
Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site
Cryo-electron microscopy structures of human RAD51 in complex with the nucleosome show that RAD51 can adopt two conformations—rings and filaments—and reveal how RAD51 binds to the nucleosome through its N-terminal lobe domain.
- Takuro Shioi
- , Suguru Hatazawa
- & Hitoshi Kurumizaka
-
Article |
 Structure of Tetrahymena telomerase-bound CST with polymerase α-primase
Structure of Tetrahymena telomerase-bound CST with polymerase α-primase
Cryo-electron microscopy structures of Tetrahymena thermophila telomerase-bound Ctc1–Stn1–Ten1 and DNA polymerase α–primase provide insights into the molecular mechanisms underlying telomere replication and maintenance.
- Yao He
- , He Song
- & Juli Feigon
-
Article |
 DNA-driven condensation assembles the meiotic DNA break machinery
DNA-driven condensation assembles the meiotic DNA break machinery
During meiosis, Mer2 and the Rec114–Mei4 complex form condensates that facilitate the formation of double-strand DNA breaks by recruiting the Spo11 transesterase complex.
- Corentin Claeys Bouuaert
- , Stephen Pu
- & Scott Keeney
-
Article |
 The interaction landscape between transcription factors and the nucleosome
The interaction landscape between transcription factors and the nucleosome
A method for systematically exploring interactions between the nucleosome and transcription factors identifies five major interaction patterns.
- Fangjie Zhu
- , Lucas Farnung
- & Jussi Taipale
-
Article |
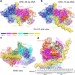 Structure of the origin recognition complex bound to DNA replication origin
Structure of the origin recognition complex bound to DNA replication origin
The cryo-EM structure of the yeast origin recognition complex (ORC) bound to a 72-base-pair origin DNA sequence provides insights into the basis of the origin selection mechanism.
- Ningning Li
- , Wai Hei Lam
- & Bik-Kwoon Tye
-
Letter |
 Alternative evolutionary histories in the sequence space of an ancient protein
Alternative evolutionary histories in the sequence space of an ancient protein
Combining ancestral protein reconstruction with deep mutational scanning to characterize alternative histories in the sequence space around an ancient transcription factor reveals hundreds of alternative protein sequences that use diverse biochemical mechanisms to perform the derived function at least as well as the historical outcome.
- Tyler N. Starr
- , Lora K. Picton
- & Joseph W. Thornton
-
Letter |
 How type II CRISPR–Cas establish immunity through Cas1–Cas2-mediated spacer integration
How type II CRISPR–Cas establish immunity through Cas1–Cas2-mediated spacer integration
Several structures of the Enteroccocus faecalis Cas1–Cas2 proteins are solved, and help to define the spacer integration mechanisms of type II CRISPR systems.
- Yibei Xiao
- , Sherwin Ng
- & Ailong Ke
-
Letter |
 Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage
Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage
CRISPR/Cas9 DNA editing creates a double-stranded break in the target DNA, which can frequently generate random insertion or deletion of bases (indels); a new genome editing approach combining Cas9 with a cytidine deaminase is described here, which corrects point mutations more efficiently than canonical Cas9, while avoiding double-stranded breaks and indel formation.
- Alexis C. Komor
- , Yongjoo B. Kim
- & David R. Liu
-
Letter |
 Crystal structure of the Rous sarcoma virus intasome
Crystal structure of the Rous sarcoma virus intasome
A crystal structure of the octameric integrase from Rous sarcoma virus in complex with viral and target DNAs.
- Zhiqi Yin
- , Ke Shi
- & Hideki Aihara
-
Article |
 The mechanism of DNA replication termination in vertebrates
The mechanism of DNA replication termination in vertebrates
This study describes a new model of eukaryotic replication termination in which converging leading strands pass each other unhindered and the replicative DNA helicase is unloaded late, after all strands have been ligated.
- James M. Dewar
- , Magda Budzowska
- & Johannes C. Walter
-
Article |
 Crystal structure of the eukaryotic origin recognition complex
Crystal structure of the eukaryotic origin recognition complex
The crystal structure of the heterohexameric origin recognition complex (ORC), essential for coordinating DNA replication onset in eukaryotes, is resolved at 3.5 Å resolution.
- Franziska Bleichert
- , Michael R. Botchan
- & James M. Berger
-
Research Highlights |
Protein crawls on chromosome
-
Article |
 DNA-binding factors shape the mouse methylome at distal regulatory regions
DNA-binding factors shape the mouse methylome at distal regulatory regions
Base-pair-resolution genomic maps of DNA methylation are generated in the mouse, providing new insights in gene regulation.
- Michael B. Stadler
- , Rabih Murr
- & Dirk Schübeler
-
Article |
 CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing
CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing
- Sanjeev Shukla
- , Ersen Kavak
- & Shalini Oberdoerffer
-
-
Letter |
 The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes
The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes
- Tian-Peng Gu
- , Fan Guo
- & Guo-Liang Xu
-
Research Highlights |
 Neuroscience: Guide the way to nerve repair
Neuroscience: Guide the way to nerve repair
-
Letter |
 Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint
Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint
Cell cycle checkpoints, such as the S-phase checkpoint, delay cell division to give the cell time to repair any damaged DNA. Here it is shown that the MLL gene — frequently disrupted in leukaemia — is part of the S-phase checkpoint. When DNA is damaged, MLL is phosphorylated by the ATR protein, causing MLL to accumulate on chromatin and methylate histone H3 on lysine 4. This delays DNA replication. MLL translocations, such as those that occur in leukaemia, disrupt this pathway and cause genomic instability.
- Han Liu
- , Shugaku Takeda
- & James J.-D. Hsieh
-
Brief Communications Arising |
 Can controversies be put to REST?
Can controversies be put to REST?
- Helle F. Jørgensen
- & Amanda G. Fisher
-
Letter |
 DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2
DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2
When double-strand breaks occur in DNA, the broken ends must undergo processing to prepare them for repair. Here, and in an accompanying study, this processing reaction has now been replicated in vitro using yeast proteins. Processing minimally requires the activities of a helicase, a nuclease and a single-strand-binding protein, although the reaction is enhanced by the addition of three factors that help to target the core complex and stimulate the unwinding activity.
- Petr Cejka
- , Elda Cannavo
- & Stephen C. Kowalczykowski
-
Letter |
 Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification
Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification
TET1 is an enzyme that catalyses the conversion of 5-methylcytosine of DNA to 5-hydroxymethylcytosine, raising the possibility that it is involved in mediating DNA demethylation. These authors show that Tet1 is involved in mouse embryonic stem cell maintenance and specification of the inner cell mass. It is required to maintain both the expression of Nanog in embryonic stem cells and the Nanog promoter in a hypomethylated state, supporting a role for Tet1 in regulating DNA methylation.
- Shinsuke Ito
- , Ana C. D’Alessio
- & Yi Zhang
-
Letter |
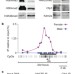 CpG islands influence chromatin structure via the CpG-binding protein Cfp1
CpG islands influence chromatin structure via the CpG-binding protein Cfp1
Most human gene promoters are embedded within CpG islands that lack DNA methylation and coincide with sites at which histone H3 lysine 4 is trimethylated (H3K4me3 sites). Here, a zinc-finger protein, Cfp1, is found to be associated with non-methylated CpG islands and H3K4me3 sites throughout the genome in the mouse brain. A primary function of non-methylated CpG islands might be to genetically determine the local chromatin modification state by interaction with Cfp1 and perhaps other CpG-binding proteins.
- John P. Thomson
- , Peter J. Skene
- & Adrian Bird
-
Letter |
 Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends
Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends
Most agents that generate breaks in DNA leave 'dirty ends' that cannot be joined immediately; instead, intervening steps are required to restore the integrity of nucleotides at the break. Here it is shown that the non-homologous end joining pathway requires a 5′-dRP/AP lyase activity to remove abasic sites at double-strand breaks. Surprisingly, this activity is catalysed by the Ku70 protein, which, together with its partner Ku86, had been thought only to recognize broken DNA ends and to recruit other factors that process ends.
- Steven A. Roberts
- , Natasha Strande
- & Dale A. Ramsden
-
Letter |
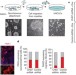 CHD7 cooperates with PBAF to control multipotent neural crest formation
CHD7 cooperates with PBAF to control multipotent neural crest formation
Heterozygous mutations in the gene encoding CHD7, an ATP-dependent chromatin-remodelling protein, result in CHARGE syndrome — a disorder characterized by malformations of the craniofacial structures, peripheral nervous system, ears, eyes and heart. In humans and Xenopus, CHD7 is now shown to be essential for the formation of multipotent migratory neural crest and for activating the transcriptional circuitry of the neural crest; shedding light on the pathoembryology of CHARGE syndrome.
- Ruchi Bajpai
- , Denise A. Chen
- & Joanna Wysocka

 Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity
Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity ATM controls meiotic double-strand-break formation
ATM controls meiotic double-strand-break formation