Featured
-
-
Article |
 The kinetic landscape of an RNA-binding protein in cells
The kinetic landscape of an RNA-binding protein in cells
Time-resolved RNA–protein cross-linking with a pulsed femtosecond ultraviolet laser, followed by immunoprecipitation and high-throughput sequencing, enables the determination of binding and dissociation kinetics of the RNA-binding protein DAZL within cells.
- Deepak Sharma
- , Leah L. Zagore
- & Eckhard Jankowsky
-
Article |
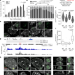 A protein assembly mediates Xist localization and gene silencing
A protein assembly mediates Xist localization and gene silencing
A protein condensate formed by multivalent interactions between the long non-coding RNA Xist and specific RNA-binding proteins drives the compartmentalization required to perpetuate gene silencing on the inactive X chromosome.
- Amy Pandya-Jones
- , Yolanda Markaki
- & Kathrin Plath
-
Article |
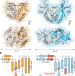 STING cyclic dinucleotide sensing originated in bacteria
STING cyclic dinucleotide sensing originated in bacteria
Structures of prokaryotic homologues of STING permit the reconstruction of the evolutionary trajectory of its incorporation into metazoan innate immunity, and reveal a role for the conserved cGAS–STING pathway in prokaryotic defence against bacteriophages.
- Benjamin R. Morehouse
- , Apurva A. Govande
- & Philip J. Kranzusch
-
Article |
 A unified mechanism for intron and exon definition and back-splicing
A unified mechanism for intron and exon definition and back-splicing
The cryo-electron microscopy structures of an early spliceosome complex in yeast reveal a unified mechanism for defining introns and exons and also for back-splicing to generate circular RNA.
- Xueni Li
- , Shiheng Liu
- & Rui Zhao
-
Letter |
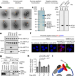 mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis
mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis
METTL3, the enzyme responsible for m6A modification, influences translation by interacting with eIF3h to mediate looping between the regions near the stop codon and 5′ cap of mRNA.
- Junho Choe
- , Shuibin Lin
- & Richard I. Gregory
-
Letter |
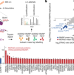 The NORAD lncRNA assembles a topoisomerase complex critical for genome stability
The NORAD lncRNA assembles a topoisomerase complex critical for genome stability
The long non-coding RNA NORAD interacts with proteins involved in DNA replication and repair, and controls the ability of RBMX to form a ribonucleoprotein complex that helps to maintain genomic stability.
- Mathias Munschauer
- , Celina T. Nguyen
- & Eric S. Lander
-
Letter |
 Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex
Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex
The assembly of single Drosophila RNA-induced silencing complexes (RISCs) is reconstituted using seven purified proteins, revealing that chaperones help stabilize the interaction of the protein heterodimer Dicer-2–R2D2 bound to the short interfering RNA with Ago2.
- Shintaro Iwasaki
- , Hiroshi M. Sasaki
- & Yukihide Tomari
-
Letter |
 Structural basis for the assembly of the Sxl–Unr translation regulatory complex
Structural basis for the assembly of the Sxl–Unr translation regulatory complex
The crystal structure of the RNA binding domains of Sxl and Unr with msl2 RNA shows that interwoven interactions establish cooperative assembly of the ternary complex, highlighting how binding of relatively general RNA binding domains to RNA can result in a unique and specific protein–RNA architecture.
- Janosch Hennig
- , Cristina Militti
- & Michael Sattler
-
News & Views |
Signalling pathways of fragile X syndrome
The RNA-binding protein FMR1 has a key role in the neurodevelopmental disorder fragile X syndrome, but the RNAs targeted by the protein were mostly unknown. An analysis reveals thousands of possible targets. See Article p.382
- Sabarinath Jayaseelan
- & Scott A. Tenenbaum
-
Article |
 FMRP targets distinct mRNA sequence elements to regulate protein expression
FMRP targets distinct mRNA sequence elements to regulate protein expression
RNA-recognition elements are identified for the fragile-X-syndrome-associated RNA-binding protein FMRP, in addition to its target messenger RNAs; although many of FMRP gene targets discovered are involved in brain function and autism spectrum disorder, a proportion are also dysregulated in mouse ovaries, suggesting cross-regulation of signalling pathways in different tissues.
- Manuel Ascano
- , Neelanjan Mukherjee
- & Thomas Tuschl
-
Letter |
 Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis
Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis
- Adelaine K. W. Leung
- , Kiyoshi Nagai
- & Jade Li
-
Letter |
 Structural basis for site-specific ribose methylation by box C/D RNA protein complexes
Structural basis for site-specific ribose methylation by box C/D RNA protein complexes
A complex of RNA and protein known as the box C/D RNP catalyses the site-specific modification of RNAs with a 2′-O-methylation group. The structure of the full complex has now been solved, including the guide RNA and either of two substrate RNAs. This structure reveals how the guide and target RNAs are aligned, and how the methyltransferase subunit, fibrillarin, facilitates placement of the target ribose into the active site.
- Jinzhong Lin
- , Shaomei Lai
- & Keqiong Ye
-
Article |
 U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation
U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation
Splicing is carried out by a collection of protein–RNA complexes known as snRNPs. The spliceosome contains equal quantities of the U1, U2, U4, U5 and U6 snRNPs, but the U1 snRNP is made in levels excess to the amounts needed to form spliceosomes, leading to the idea that excess U1s might have splicing independent functions. Here it is shown that the U1 snRNA interacts with some pre mRNAs whose introns have cryptic polyadenylation sites. This interaction prevents premature termination and polyadenylation of the pre mRNA.
- Daisuke Kaida
- , Michael G. Berg
- & Gideon Dreyfuss
-
Letter |
 Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2
Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2
The association of microRNAs with Argonaute proteins (AGOs) yields complexes regulating gene expression. Although bacterial and archaeal miRNAs show no sequence preference at their 5′ ends, eukaryotic miRNAs tend to have a 5′ U or A. Here the structure of the human AGO2 MID domain complexed with ribonucleotide monophosphates is solved, revealing specific interaction of UMP and AMP with a loop that discriminates against CMP or GMP, and explaining the observed preference.
- Filipp Frank
- , Nahum Sonenberg
- & Bhushan Nagar

 Structure of active human telomerase with telomere shelterin protein TPP1
Structure of active human telomerase with telomere shelterin protein TPP1