Abstract
We have discovered two types of 5′ intronic gene mutation that impair androgen receptor (AR) mRNA expression severely, and cause complete androgen insensitivity. Labium majus skin fibroblasts (LMSF) hemizygous for each mutation had negligible specific androgen binding, and did not react to an antibody against an N-terminal peptide of the AR. Both mutations were detected by direct sequencing of exons PCR-amplified with flanking primers. One mutation is an adenine to thymine transversion at position +3 of the intron 6 splice-donor site. Using LMSF mRNA, RT-PCR of a portion of the AR androgen-binding domain yielded a small amount of a 302-bp mutant fragment instead of a 433-bp wild-type product. Sequencing established that exon 5 was followed, out of frame, by exon 7: exon 6 was skipped. The other mutation is a thymine insertion at the +3 position of the intron 1 donor-splice site. RT-PCR and sequencing revealed a small amount of normal-size mRNA with normal exon 1-exon 2 splicing. Quantitative RT-PCR on mutant LMSF showed AR mRNA levels were well below 10% of normal; hence, most of the aberrant AR mRNA resulting from each mutation is probably unstable. The misbehavior caused by these two mutations indicates that in the AR the splice-donor site +3 adenine is critical; indeed, 57% of eukaryotic introns have adenine in the +3 position, while only 2% have thymine.
Similar content being viewed by others
Introduction
The androgen receptor is a polymorphic (910–919 amino acids), androgen-activated, DNA-binding protein that regulates the transcription of certain target genes [1]. The Xq11–12-linked androgen receptor gene (AR) and its product have the typical intron-exon composition and structure-function organization of other members in the steroid receptor family (fig. 1). In hemizygotes a large variety of AR alterations yield androgen insensitivity varying in severity from a female phenotype, to various degrees of external genital ambiguity, to a male phenotype with subnormal virilization at puberty. In the AR gene mutation database [2], about 80% of the nearly 150 different AR alterations are due to single-base mutations that cause premature termination codons or amino acid substitutions within the AR protein; only a small minority (3%, including the mutations characterized herein) cause splice-junction abnormalities [1]. The three previously reported examples of AR splice-donor site mutations involve substitution of the invariant guanine at the +1 position by thymine [3] or adenine [4, 5]. In this paper we describe the discovery, and the phenotypic consequences, of two different single-base mutations that put thymine in place of adenine at the +3 position in two splice-donor sequences of the AR. The critical nature of the +3 splice-donor site position in normal mRNA processing is highlighted by the identification of 14 other +3 splice-donor site mutations in a variety of human genetic diseases [6–17, and references to +3 mutations therein].
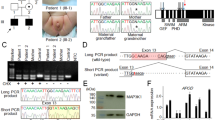
The AR gene and protein: untranslated regions (UTR), and intron-exon composition. Exon 1 encodes the transactivation modulatory domain; exons 2–3 and 4–8 form the DNA and androgen-binding domains, respectively. Alternative 3′ processing yields transcripts of ∲ 8 and 11 kb. (Gln)n and (Gly)n denote the polymorphic polyglutamine and polyglycine tracts in the AR’s N-terminal domain. Shown below are the primer combinations used for quantitative-competitive RT-PCR (solid lines) and for attempts to detect abnormal splicing in 926203 (dashed lines).
Subjects and Methods
Subjects
Subject 4479 (referred by Dr. H.N. Valentine, The University of Western Ontario, London, Ont., Canada) had a right inguinal herniorrhaphy at age 5 during which histologically identified testicular tissue was removed. She had normal female external genitalia but lacked a uterus. The peripheral lymphocyte karyotype was 46,XY. The mother of 4479 had two maternal aunts with primary amenorrhea; thus, we presumed she was an obligate heterozygote. Two skin fibroblast substrains, coded 7534 and 7534Q, were derived from a single expiant of her labium majus skin.
Subject 926203 presented at 15 years with primary amenorrhea and a height of 171.4 cm, well above the sex-adjusted genetic target height of 159 ± 8.5 cm. She had high blood levels of luteinizing hormone and testosterone and well-developed breasts, but no pubic or axillary hair. The vagina was 4 cm and blind. The peripheral lymphocyte karyotype was 46,XY. In the family history there were no maternally-related females with primary amenorrhea, delayed menarche, or sparse, delayed or asymmetrical sexual hair, and no maternally-related males with hypospadias, gynecomastia, or unexplained infertility.
Androgen Binding by Labium Majus Skin Fibroblasts (LMSF)
Replicate confluent monolayers in 6-cm Petri dishes were incubated with 3 nM [3H]mibolerone (MB) alone, in triplicate, to measure total binding, or together with 600 nM radioinert MB for 2 h at 37 °C, in duplicate, to measure nonspecific binding [18]. Specific binding was determined by subtracting nonspecific from total binding, and dividing by protein concentration as determined by the Lowry assay [19].
Genomic PCR Amplification and Direct Sequencing of PCR Products
Exons 1–8 were amplified and sequenced as described [20]. In the sequencing ladders shown (fig. 2), exon 6 and a portion of intron 6 were sequenced using an intron 6-specific primer (14S); exon 1 and a portion of intron 1 were sequenced with an intron 1-specific primer (7AS). (See table 1 for all primer sequences; positions according to Lubahn et al. [21], Genbank accession No. JO3180.)
DNA sequencing ladders showing the AR splice-donor site mutations. Lower-case letters distinguish intronic from exonic bases. A An A to T transversion was identified at position +3 of intron 6 of subject 4479 [Int6(+3A>T)]; her mother (7534) was heterozygous for the same mutation. B A T insertion was found at the same position of intron 1 in subject 926203 [Int1(+3insT)].
RT-PCR and Direct Sequencing of RT-PCR Products
Total RNA, isolated as described [22], was reverse transcribed using random hexanucleotide primers (Boehringer Mannheim) and Superscript RNase H− Reverse Transcriptase (Gibco BRL, Life Technologies, Inc.) according to the manufacturers’ directions. The RT products were PCR amplified in 100 µl volume with 100 µM each dNTP, 2 units Taq polymerase and 50 pmol each primer for 35 cycles with denaturation, annealing and elongation for 1 min each at 95, 55, and 72°C, respectively. Exon 4-specific (11S) and exon 7-specific (16AS) primers were used to amplify a segment of the 4479, 7534, and 7534Q AR mRNA encoding the androgen-binding domain, as shown in figure 3. The products were electrophoresed on a 1.5% agarose gel-containing ethidium bromide, photographed, and compared to size standards. In the sequencing ladders shown (fig. 4), RT-PCR products from 4479 and 7534 were amplified using 11S and 18AS then sequenced using an exon 7-specific primer (17AS). Polyglutamine from 4479 [Int6(+3A>T)] lacked exon 6. The consequent frameshift results in a premature termination codon in exon 7. (X)8 = Eight unspecified amino acids. (CAG)n and polyglycine (GGN)n lengths were determined as described [20].
Total RNA from 2080 (normal), 4479, 7534 and 7534Q fibroblasts was reverse-transcribed and PCR-amplified (RT-PCR) using exon 4- and exon 7-specific primers (1 IS and 16AS). The products were analyzed on a 1.5% low-melt agarose gel with HaeIII-digested ΦX174RF markers (ΦX). The normal product expected is 433 bp; shorter fragments (302 bp) were observed for 4479 [Int6(+3A>T)], and 7534Q.
Competitive-quantitative RT-PCR using total RNA from 926203 or 4479 LMSF (fig. 6) was performed as described [20] except 35 PCR cycles were completed using exon 4–7-specific primers 11S and 16AS. The reactions containing 4479 products were not digested, but the normal and 4479 fragments were easily distinguishable by size. Exon 1-specific competitive-quantitative RT-PCR was carried out essentially as for exons 4–7 except primers 1S and 3AS were used. The reactions included DMSO (2% final volume) and a competitor plasmid with a Bsr F1 site due to a silent C → G mutation at codon 390 of the AR [D.M. Vasiliou, MSc thesis, McGill University]. We performed 35 cycles at 98°C for 1 min, 55 °C for 1 min and 75 °C for 1.5 min. After digestion with Bsr Fl, samples were electrophoresed and quantitated as described [20].
Quantitative-competitive RT-PCR. A Exon 4-7-specific primers 11S and 16AS were used to determine the level of AR mRNA in 926203 fibroblasts relative to normal 2080. B The ethidium bromide-stained gel containing digested PCR products whose negative was scanned to obtain the results plotted in A. M = HaeIII-ΦX174 markers; lanes 1–9 = the dilution series of the competitor AR cDNA ranged from 400 to 1.6 pg/reaction for 2080 and from 50 to 0.2 pg/reaction for 926203. C The ethidium bromide-stained gel showing undigested exon 4–7 products of reverse transcribed 4479 fibroblast mRNA (sample) and the same plasmid competitor AR cDNA used above (competitor). The competitor ranged from 50 to 0.4 pg/reaction (lanes 1–8). No competitor was included in lane 9. HaeIII-ΦX174 marker sizes are indicated.
Total cell lysates from 926203, 7534Q (one of two substrains from the mother of 4479), 4479, 8812, 2200 and 2080 fibroblasts (350 µg protein each) were electrophoresed on a 7% SDS-PAGE gel. AR genotype is indicated below: Intl(+3insT) and Int6(+3A>T) = +3 splice-donor site mutations; AR del = complete deletion of AR; + = normal. The blot was probed with (A) an AR-specific monoclonal antibody F39.4.1, then (B) a 70-kD heat shock protein (hsp)-specific monoclonal antibody to control for amount of protein loaded. The blot was developed using the ECL Western blotting chemiluminescence detection system (Amersham). The sizes of the Rainbow protein molecular weight markers (Amersham) are shown.
Western Blotting
Extracts from the LMSF of 926203, 7534Q, 4479, and controls were prepared as described [18] and 350-µg protein samples subjected to electrophoresis on a 7% SDS-PAGE gel. After electroblotting, the nitrocellulose filter was blocked in 5% skim milk powder in 0.5% Tween/10 mM Tris HCl pH 7.5, 150 mM NaCl (Tris-buffered saline (TBS)) and processed as described [25] using the anti-AR monoclonal antibody F39.4.1 [26]. The filter was then washed in 0.5% Tween/TBS, reacted with a monoclonal antibody against constitutively expressed hsp 70 (Stressgen Biotechnologies Corporation), diluted 1:1,000 in 0.5% Tween/TBS, and processed as above.
Splice-Donor Site Analysis
The consensus values (CV) for each normal and mutant splice-donor site were calculated according to the method of Shapiro and Senapathy [25] using primate values for nucleotide percentages at each of eight splice-donor site positions. CV = 100 (t-mint)/(maxt-mint), where t = the total of the percentages for the eight positions of a single site and mint and maxt are the minimum and maximum percentage totals possible for primate splice-donor sites.
Results
Androgen Binding and AR Sequence Alterations in LMSF
We measured androgen-binding activity in LMSF of subjects 4479 and 926203, and in two LMSF substrains (7534 and 7534Q) derived from the mother of 4479. The cells of 4479 and 926203 had negligible androgen-binding activity. However, one of the LMSF substrains derived from the mother of 4479 had very low specific androgen binding (2–5 fmol/mg protein; strain 7534Q), while the other substrain had normal androgen binding (22 fmol/mg protein, strain 7534; normal range 15–40 fmol/mg protein). These results indicated cell mosaicism due to differential X-chromosome inactivation, and strongly supported her presumptive diagnosis as an obligate heterozygote.
To determine if sequence alterations in the androgen-binding domain (ABD) of the AR were the cause of the negligible androgen binding in subjects 4479 and 926203, exons 4–8 were PCR amplified from LMSF-derived genomic DNA and the sequences of the exons and exon/intron boundaries were determined. The only alteration found in subject 4479 was an A to T transversion in the +3 position of intron 6 [Int6(+3A > T)], as shown in figure 2A. Coexistent mutations in and around exons 2 and 3 were excluded. Genomic sequencing (fig. 2A) proved that 7534, the mother of 4479, was heterozygous for Int6(+3A > T), as predicted by the androgen-binding assays. As no mutation was found in or around exons 2–8 of subject 926203, direct sequencing was extended to the translated portion of exon 1 and its flanking sequences. Figure 2B shows the only mutation found: a T insertion at the +3 position of intron 1 [Int 1(+3insT)]. This mutation was not present in the subject’s mother, as predicted from her family history.
RT-PCR Analysis of AR Transcripts
Total RNA from 2080 (normal), 4479, 7534 and 7534Q fibroblasts was reverse transcribed (RT) and the cDNA subjected to PCR amplification using exon 4- and exon 7-specific primers within the ABD. Figure 3 shows that the normal sample yields the expected 433-bp fragment, as does substrain 7534, while a shorter one (302 bp) is observed clearly for 4479, and for substrain 7534Q from her heterozygous mother. Sequencing of a PCR product that included the 302-bp fragment shown in figure 3 revealed that the 131-bp exon 6 is missing (fig. 4); thereby fully accounting for the reduced size of the mutant fragment (fig. 3). Furthermore, skipping of exon 6 with consequent exon 5-exon 7 fusion creates a frameshift that would result in 12 altered amino acids followed by a premature stop codon (TGA). The predicted truncated AR would have 783 amino acids, a molecular weight of 82.6 kDa, and lack the portion of the ABD encoded by exons 6–8.
The normal and mutant fragments in substrains 7534 and 7534Q, respectively, indicated differential X-chromosome inactivation. This was confirmed by sequencing the glutamine [Gln;(CAG)n] tracts of exon 1-specific RTPCR products from 4479, 7534 and 7534Q. The 7534 product had 28 Gln codons, while the 7534Q product had 21 Gln codons. The latter corresponded to the 21 Gln-codon tract of the AR on the single X chromosome of 4479.
All the primer combinations used for RT-PCR of exon 1 and exons 2–8 of 926203’s AR mRNA produced greatly reduced quantities of normal-size fragments. Competitive RT-PCR using the primers identified in figure 1 yielded a value of 6% of normal for exon 1 (results not shown) and 5% of normal for exons 4–7 (figs. 5A, B). Multiple primer pairs that spanned the exon 1/exon 2 junction were used in an attempt to isolate 926203 AR mRNA splicing variants. No products were seen however, except on one occasion with a combination of primers 2S and 8AS, when sequencing showed the exon 1/exon 2 junction was normal, and the authenticity of the RT-PCR product was confirmed by sequencing the polyglycine (GGN)n tract. Both the RT-PCR product and 926203 genomic DNA contained 23 glycine codons. Northern analysis of poly A+ mRNA from 926203 GSF also did not reveal any AR mRNA variants (results not shown).
AR mRNA expression in 4479 was found to be very low (fig. 5C). In a separate quantitative RT-PCR experiment, using 2080 RT-PCR product rather than plasmid DNA for competition, and no restriction enzyme digestion, the level of the mutant transcript in 4479 was 4% of normal AR mRNA (results not shown).
Western Blot Analysis
To determine if AR proteins were produced by LMSF from 926203, 4479 and 7534Q, cell extracts were electrophoresed, transferred to a nitrocellulose filter and reacted with an anti-hAR antibody (F39.4.1 [24]). No AR protein was detectable, either of normal size in 926203, or of reduced size in 4479 or 7534Q (fig. 6). Probing with an hsp 70 antibody showed equivalent protein loading for all samples.
Splice-Donor Site Analysis
All 7 splice-donor sites in the AR were analyzed by the method of Shapiro and Senapathy [25]. The CV, or homology score, is a measure of the similarity of a given sequence to the 5′ AG:GUA/GAGU 3′ consensus sequence for splice-donor sites. At a given site the CV can be between 0 and 100; most normally used splice sites have a CV of >70 [8]. The CV values in the AR range from 94.3 for intron 3 to 73.2 for intron 7. The Intl(+3insT) mutation drops the CV of intron 1 dramatically, from 91.8 to 62.8, because the T insertion shifts the bases out of alignment with the consensus (table 2). The Int6(+3A > T) mutation drops the intron 6 CV from 79.7 to 69.7, a lower score than that for any normal splice-donor site in the AR or splice-donor sites in general.
Discussion
Cooper and Krawczak [26] estimated that point mutations altering mRNA splicing represent ∼ 15% of all those causing human genetic disease, and splice-donor sites are more often affected than splice-acceptor sites [27]. As of October 1996, only 6 (including the 2 reported here) of the 143 (4.2%) point mutations or small deletions or insertions associated with androgen insensitivity in the AR Gene Mutations Database [2] affect splicing; and, appropriately, 5 of the 6 are at splice-donor sites. Despite their infrequency, the analysis of natural splicing mutations can illuminate basic mechanisms of mRNA transcription and processing. Three of the 4 AR splice mutations reported previously [3–5] caused complete androgen insensitivity by substitution at the invariant +1 G position of the intron 3, 4, or 7 splice-donor sites. In the fourth, deletion of the intron 2 branch-point sequence caused partial androgen insensitivity [28]. The 2 mutations described here affirm that alterations at the +3 position of a splice-donor site disrupt normal AR mRNA processing as much as alterations at the +1 position. Splice-donor site +3 mutations have been incriminated as the cause of human genetic disease in 14 other cases [6–17 and references therein]. Of the total, 3 are +3 A > T [8, 12, 17], as in our subject 4479, and 4 are +3insT [6, 9, 11, 14], as in our subject 926203.
The phenotypic consequence of the AR Int1(+3insT) or the Int6(+3A > T) mutation is complete androgen insensitivity. LMSF from both hemizygous subjects, and from one of two Int6(A > T) heterozygous LMSF substrains (7534Q) had little androgen-binding activity and no AR protein detectable by Western blotting. AR mRNA was expressed at levels less than 10% of normal in both cases, indicating that mRNA processing is adversely affected by both +3 splice-donor site mutations. Analysis of RT-PCR products from 4479 [Int6(A > T)] demonstrated that mutant mRNA was produced; exon 6 was skipped. Exon skipping was also observed in the 7534Q substrain but not in the 7534 substrain. This may be explained by the chance development of two different LMSF subpopulations with differential X-chromosome inactivation. The active X chromosome in the 7534 cells must bear the normal allele and thus produce normal AR protein capable of binding androgen, while the 7534Q cells express the mutant allele, as confirmed by determination of polyglutamine-codon tract lengths in RT-PCR products.
We were not able to demonstrate aberrant splicing in LMSF of 926203 [Int1(+3insT)], despite the fact that splice-donor mutations can cause cryptic splice-site utilization or intron retention in addition to exon skipping [29]. Presumably exon 1 skipping does not occur in strain 926203 because there is no upstream slice-donor site to which the splice-acceptor site of exon 2 can be joined. In the single other case with a +3 splice-donor site mutation in intron 1, two mutant transcripts utilizing a cryptic splice site 20 bp into intron 1 were detected [14]. Atypically, in one of these transcripts, skipping of exon 2, but not of the exon upstream of the mutation (exon 1), occurred. With the exception of one pair of primers whose product contained a normal junction, PCR amplification with multiple primer pairs spanning the exon1/exon 2 boundary of 926203 did not generate any DNA fragments. This indicates that cryptic splice sites in the immediate vicinity of the normal splice-donor site of exon 1 in 926203 were not used. This also suggests that the low level AR mRNA measured by competitive RT-PCR in 926203 was probably spliced normally. Retention of intron 1 would have resulted in addition of >24 kb to the already lengthy AR message (∼ 8 or 11 kb), but Northern analysis of 926203 poly A+ RNA also failed to reveal any longer than normal AR mRNA species. Possibly, long-range PCR would be a more sensitive technique for revealing abnormal splicing patterns in 926203.
Base changes that alter the splice-donor site consensus sequence severely could hinder base-pairing between the 3′ UCCAUUCA 5′ sequence at the 5′ end of U1 snRNA and the 5′ AG:GUA/GAGU 3′ consensus sequence of a mammalian splice-donor site and therefore affect spliceosome assembly [30]. The consensus values calculated for the mutations described here indicate serious alteration of the consensus sequence at the exon-intron borders 1 and 6. The consequence for Int1(+3insT) is a markedly reduced amount of normal mRNA; that for Int6(+3A > T), a markedly reduced amount of aberrant mRNA, reinforces the exon definition mode of splice-site selection [31]. Exon skipping results when the splice-donor site of the previous exon is joined to the splice-acceptor site of the following exon; this was demonstrated clearly for the Int6(+3A > T) mutation. Classical exon skipping also occurs with 12 of the 14 intron +3 mutations previously recognized as causing human genetic disease; it was not sought in the 14th case [11].
As a consequence of exon skipping, the Int6(+3A > T) mutation yields a mutant transcript that is predicted to generate a premature stop codon (PSC) in the proximal portion of exon 7, the penultimate exon of the AR. Such PSCs are usually associated with a reduced level of mRNA, rather than production of a truncated polypeptide. This is explainable by the ‘translational translocation’ or ‘nuclear scanning’ models that link normal nuclear-cytoplasmic transport of mRNA to the integrity of mRNA translation [32]. Indeed, destabilization of the full-length mRNA due to premature termination of translation has been reported for human β-globin trans-genes containing nonsense codons [33]. In any event, any protein translated from the mutant transcript would be unable to bind androgen, and it is not sufficiently stable to be detectable by Western blotting. Likewise, the low amount of normal AR mRNA found in 926203 isinsufficient to support any male sexual morphogenesis.
In conclusion, the abnormal mRNA expression caused by the two AR mutations reported herein demonstrates the critical role that the +3 position of a splice-donor site can play in mRNA processing and development of human disease.
References
Quigley CA, De Bellis A, Marschke KB, El-Awady MK, Wilson EM, French FS: Androgen receptor defects: Historical, clinical, and molecular perspectives. Endocr Rev 1995; 16:271–321.
Gottlieb B, Trifiro M, Lumbroso R, Pinsky L: The androgen receptor gene mutations database. Nucleic Acids Res 1997;25:158–162.
Ris-Stalpers C, Kuiper GGJM, Faber PW, Schweikert HU, van Rooij HCJ, Zegers ND, Hodgins MB, Degenhart HJ, Trapman J, Brinkmann AO: Aberrant splicing of androgen receptor mRNA results in synthesis of a nonfunctional receptor protein in a patient with androgen insensitivity. Proc Natl Acad Sci USA 1990;87:7866–7870.
Evans BAJ, Ismail RA, France T, Hughes IA: Analysis of the androgen receptor gene structure in a patient with complete androgen insensitivity syndrome. J Endocrinol 1991:129 (suppl):abstr 65.
Yong EL, Roy A, Chua KL, Ratnam S, Yang M: Complete androgen insensitivity due to a splice-site mutation in the androgen receptor gene and genetic screening with single-stranded conformation polymorphism. Fertil Steril 1994;61:856–862.
Hidaka Y, Palella TD, O’Toole TE, Tarlé SA, Kelley WN: Human adenine phosphoribosyl-transferase: Identification of allelic mutations at the nucleotide level as a cause of complete deficiency of the enzyme. J Clin Invest 1987; 80:1409–1415.
Carstens RP, Fenton WA, Rosenberg LR: Identification of RNA splicing errors resulting in human ornithine transcarbamylase deficiency. Am J Hum Genet 1991;48:1105–1114.
De Boer M, Bolscher BGJM, Dinauer MC, Orkin SH, Smith CIE, Ahlin A, Weening RS, Roos D: Splice site mutations are a common cause of X-linked chronic granulomatous disease. Blood 1992;80:1553–1558.
Sankila EM, Tolvanen R, van den Hurk JAJM, Cremers FPM, de la Chapelle A: Aberrant splicing of the CHM gene is a significant cause of choroideremia. Nat Genet 1992;1:109–113.
Bidichandani SI, Shiach CR, Lanyon WG, Connor JM: A novel splice donor mutation affecting position +3 in intron 6 of the factor VIII gene. Hum Mol Genet 1994;3:651–653.
Inazu A, Jiang XC, Haraki T, Yagi K, Kamon N, Koizumi J, Mabuchi H, Takeda R, Takata K, Moriyama Y, Doi M, Tall A: Genetic cholesteryl ester transfer protein deficiency caused by two prevalent mutations as a major determinant of increased levels of high density lipoprotein cholesterol. J Clin Invest 1994;94:1872–1882.
Kaler SG, Gallo LK, Proud VK, Percy AK, Mark Y, Segal NA, Goldstein DS, Homes CS, Gahl WA: Occipital horn syndrome and a mild Menkes phenotype associated with splice site mutations at the MNK locus. Nat Genet 1994; 8:195–202.
Mertes G, Ludwig M, Finkelnburg B, Krawczak M, Schwaab R, Brackmann HH, Olek K: A G+3-to-T donor splice site mutation leads to skipping of exon 50 in von Willebrand factor mRNA. Genomics 1994;24:190–191.
Morrone A, Morreau H, Zhou XY, Zammarchi E, Kleijer WJ, Galjaard H, dAzzo A: Insertion of a T next to the donor splice site of intron 1 causes aberrantly spliced mRNA in a case of infantile GM1-gangliosidosis. Hum Mutat 1994;3:112–120.
Purandare SM, Lanyon WG, Connor JM: Characterisation of inherited and sporadic mutations in neurofibromatosis type-1. Hum Mol Genet 1994;3:1109–1115.
Brackett JC, Sims HF, Rinaldo P, Shapiro S, Powell CK, Bennett MJ, Strauss AW: Two alpha subunit donor splice site mutations cause human trifunctional protein deficiency. J Clin Invest 1995;95:2076–2082.
Miyaki M, Konishi M, Muraoka M, Kikuchi-Yanoshita R, Tanaka K, Iwama T, Mori T, Koike M, Ushio K, Chiba M, Nomizu S, Utsunomiya J: Germ line mutations of hMSH2 and hMLH1 genes in Japanese families with hereditary nonpolypopsis colorectal cancer (HNPCC): Usefulness of DNA analysis for screening and diagnosis of HNPCC patients. J Mol Med 1995;73:515–520.
Shkolny DL, Brown TR, Punnett HH, Kaufman M, Trifiro MA, Pinsky L: Characterization of alternative amino acid substitutions at arginine 830 of the androgen receptor that cause complete androgen insensitivity in three families. Hum Mol Genet 1995;4:515–521.
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ: Protein measurement with the Folin phenol reagent. J Biol Chem 1951;193:265–275.
Beitel LK, Prior L, Vasiliou DM, Gottlieb B, Kaufman M, Lumbroso R, Alvarado C, McGillivray B, Trifiro M, Pinsky L: Complete androgen insensitivity due to mutations in the probable alpha-helical segments of the DNA-binding domain in the human androgen receptor. Hum Mol Genet 1994;3:21–27.
Lubahn DB, Joseph DR, Sar M, Tan J, Higgs HN, Larson RE, French FS, Wilson EM: The human androgen receptor: Complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol Endocrinol 1988;2:1265–1275.
Trifiro M, Prior RL, Sabbaghian N, Pinsky L, Kaufman M, Nylen EG, Belsham DD, Greenberg CR, Wrogemann K: An amber mutation creates a diagnostic MaeI site in the androgen receptor gene of family with complete androgen insensitivity. Am J Hum Genet 1991;40: 493–499.
Kazemi-Esfarjani P, Beitel LK, Trifiro M, Kaufman M, Rennie P, Sheppard P, Matusik R, Pinsky L: Substitution of valine-865 by methionine or leucine in the human androgen receptor causes complete or partial androgen insensitivity, respectively with distinct androgen receptor phenotypes. Mol Endocrinol 1993;7:37–46.
Zegers ND, Ciaassen E, Neelan C, Mulder E, van Laar JH, Voorhorst MM, Berrevoets CA, Brinkmann AO, van der Kwast TH, Ruizeveld de Winter JA, Trapman J, Boersma WJA: Epitope prediction and confirmation for the human androgen receptor: Generation of monoclonal antibodies for multi-assay performance following the synthetic peptide strategy. Biochim Biophys Acta 1991;1073:23–32.
Shapiro MB, Senapathy P: RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res 1987; 15: 7155–7174.
Cooper DN, Krawczak M: Human Gene Mutation. Aransas Pass/Tex, BIOS Scientific Publishers, 1993.
Nakai K, Sakamoto H: Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene 1994; 141: 171–177.
Ris-Stalpers C, Verleun-Mooijman MC, de Blaeij TJ, Degenhart HJ, Trapman J, Brinkmann AO: Differential splicing of human androgen receptor pre-mRNA in X-linked Reifenstein syndrome, because of a deletion involving a putative branch site. Am J Hum Genet 1994;54:609–617.
Maruyama T, Miyake Y, Tajima S, Funahashi T, Matsuzawa Y, Yamamoto A: A single point mutation in the splice donor site of the low-density-lipoprotein-receptor gene produces intron read-through, exon-skipped and cryptic-site-utilized transcripts. Eur J Biochem 1995; 232:700–705.
Horowitz DS, Krainer AR: Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet 1994; 10:100–106.
Robberson BL, Cote GJ, Berget SM: Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol 1990; 10:84–94.
Mcintosh I, Hamosh A, Dietz HC: Nonsense mutations and diminished mRNA levels. Nat Genet 1993;4:219.
Lim SK, Sigmund CD, Gross KW, Maquat LE: Nonsense codons in human β-globin mRNA result in the production of mRNA degradation products. Mol Cell Biol 1992;12:1149–1161.
Acknowledgements
This work was supported by the Medical Research Council of Canada, the Cancer Research Society of Canada, the Fonds pour la Formation de Chercheurs et l’Aide à la Recherche du Québec, and the Fonds de la Recherche en Santé du Québec. We are grateful to Mary Tomaras and Rhona Rosenzweig for faithful secretarial assistance.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Trifiro, M.A., Lumbroso, R., Beitel, L.K. et al. Altered mRNA Expression due to Insertion or Substitution of Thymine at Position +3 of Two Splice-Donor Sites in the Androgen Receptor Gene. Eur J Hum Genet 5, 50–58 (1997). https://doi.org/10.1007/BF03405877
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03405877