Abstract
The meiotic segregation of chromosomes 10 and 12 was analyzed in a male heterozygous for a reciprocal translocation, t(10;12)(q26.1;p13.3), using fluorescence in situ hybridization (FISH). Centromeric specific probes that detect alpha satellite sequences of chromosomes 10 and 12 were used. A total of 10,049 spermatozoa were analyzed. The frequencies of alternate/adjacent 1, adjacent 2, and 3:1 modes of segregation were: 84.25, 10.95%, and 4.42%, respectively. Diploidy was present in 0.23% of spermatozoa. Similar segregation patterns have been reported for this donor by direct karyotyping of sperm cells. FISH is a valuable technique for studying meiotic segregation patterns in that larger samples can be studied in a relatively short time. However, it does not provide information on the full chromosome complement of the spermatozoon.
Similar content being viewed by others
Introduction
Chromosome translocations are relatively common in humans. They result from the exchange between nonhomologous autosomes. Approximately one couple out of 600 carries a reciprocal translocation [1]. Individuals who are heterozygous are at an increased risk for fetal wastage and for having children with abnormal chromosomes. A balanced carrier of a reciprocal translocation is potentially able to produce a variety of gametes by meiotic segregation. All types of segregations have been described in reciprocal translocation carriers by karyotyping pronu-clear sperm chromosomes after fusion of human sperm with zona-free hamster eggs. Data are available from 37 reciprocal translocation carriers [for reviews, see ref. 2, 3]. A literature review of sperm chromosomes data provided little evidence that different types of reciprocal translocations tend to segregate different types of imbalanced gamete preferentially. In fact, independent of the type of translocation and the expected mode of imbalance at term, adjacent 1 segregation predominates in many translocation carriers. However, there are some exceptions such as four translocations with a statistically higher number of adjacent 2 segregants [reviewed ref. 2]. These translocations all shared a common feature, namely they all had shorter combined interstitial segments, in agreement with the hypothesis of Jalbert et al. [4].
With the advent of fluorescence in situ hybridization (FISH), and the development of reliable methods to decondense sperm chromatin, larger numbers of sperm nuclei can be screened relatively rapidly. This approach is presently limited to the detection of numerical aberrations and can be applied to the study of the meiotic segregation of translocations.
This report provides FISH results on the segregation patterns of the spermatozoa of a male heterozygous for a reciprocal translocation t(10;12)(q26.1; p13.3). Sperm chromosome analysis of this translocation has already been reported [2]. The overall distribution of gamete segregation types is similar in both studies.
Materials and Methods
The translocation heterozygote who was 40 years old at the time of the study, was ascertained because of repeated reproductive failures that ended in miscarriage or termination. One pregnancy for which cytogenetic studies were available, was the result of an adjacent 1 segregation with trisomy for 12p13.3→12pter and monosomy for 10q26.1→10qter.
The fresh ejaculate was allowed to liquefy at room temperature. An aliquot of the specimen was washed twice in BWW [5] and the sperm suspension was used to make smears which were allowed to air dry and were subsequently stored at −20°C until use. The rest of the specimen was capacitated and used for sperm chromosome analysis [2].
Sperm heads were decondensed for FISH following procedures described by Wyrobek et al. [6] with minor modifications. The slides were brought up to room temperature, incubated in 1.0 mM dithiothreitol (DTT, Sigma) for 30 min, followed by a second incubation in 20 mM lithium diiodosalicylate (LIS) (Sigma) for 3 h and a brief rinse in 2 × SSC. Immediately afterwards, the slides were processed for FISH.
Probes that detect the alpha satellite sequences of chromosomes 10 and 12 were used (CEP 10 and CEP 12). Both were obtained from VYSIS. The probes had been directly labelled with the following fluorochromes: d-UTP, orange, for chromosome 10, and d-CTP, green, for chromosome 12. The sperm slides were denatured in 70% formamide/2 × SSC at 75 °C for 10 min. Ten microliters of the probe-hybridization mixture (7 µl hybridization buffer, 1 µl of H2O, and 1 µl of each probe) were denatured at 75 °C for 5 min, added to the slides and hybridized overnight at 42 °C. The hybridization buffer contained 10% dextran sulfate, 55% formamide in 1 × SSC. After posthybridization washes at 50% stringency and 45 °C, the slides were counterstained with DAPI.
The microscope analysis was performed at a Zeiss Axiophot with a triple band-pass filter for FITC, Rodamine and DAPI allowing the simultaneous visualization of orange, green and blue. Only individual and well deli-nated spermatozoa with a tail were scored. In cases where there was doubt, phase contrast microscopy was used to confirm the presence of a tail. Two signals of the same color were counted as disomy only if they were of the same size and intensity and separated at least by approximately one domain. The efficiency of hybridization was calculated by counting the number of hybridized spermatozoa in the first 500 scored sperm cells in each slide. Two microscope slides were scored for this study, with hybridization efficiencies of 98 and 99%, respectively.
Tests for equality of two binomial proportions [7] were used to determine, for example, equality of proportions of the products of adjacent 2 segregation. Tests for equality of the distribution of segregation products between FISH segregation analysis and sperm cytogenetic analysis used a contingency table analysis [7].
Results
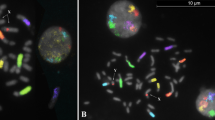
A total of 10,049 sperm nuclei from the translocation heterozygote t(10; 12)(q26.1;p 13.3) were analyzed. Examples are provided in figure 1. The segregation patterns were determined by the presence/absence of chromosomes 10 and 12. Table 1 shows a breakdown of the results. The number of spermatozoa with one copy of chromosome 10 and one copy of chromosome 12 was 8,466 (84.25%). As the normal 10 and 12 cannot be distinguished from their derivatives, alternate and adjacent 1 segregations exhibit the same hybridization patterns. A total of 1,101 (10.95%) sperm segregated to the adjacent 2 mode of segregation. For adjacent 2 segregations, the number of cells with a presumably normal 10 and a derivative 10 was 534 (5.31 %) and for a presumably normal 12 and a derivative 12 was 567 (5.54%). The ratio of the two complementary products of adjacent 2 segregation is maintained at 1:1 in this translocation heterozygote, i.e., the number of cells with a normal 10 and a presumably translocated 10 (534) and the number of cells with a normal 12 and a presumably translocated 12 (567) were not significantly different. 3:1 segregation were less frequent with a total of 444 (4.42%). 3:1 segregations should theoretically result in equal numbers of nuclei with one signal and three signals. In this study, the number of cells with one signal significantly exceeded the number of nuclei with three signals for both types of complementary products (-/12; 10/10/12, p <0.0001 and -/10; 10/12/12, p < 0.0001). Twenty-three nuclei (0.23%) showed a 10/10/12/12 diploid complement. Fifteen cells had three copies of the same chromosome (0.15%). These cells could be the result of adjacent 2 segregation plus nondisjunction at anaphase II. The results of the comparison of this FISH segregation analysis with the sperm cytogenetic analysis showed no significant differences (p = 0.5103 by contingency table analysis) in the segregation patterns of this translocation (table 2).
Spermatozoa from a male heterozygous for a t(10; 12) labeled with CEP 10 and CEP 12 (Vysis). a Two nuclei showing one red (chromosome 10) and one green signal (chromosome 12) corresponding to an alternate or adjacent 1 segregation. One sperm nucleus with a 10/12/12 pattern corresponding to a 3:1 segregation (arrow), b One diploid spermatozoa with a 10/10/12/12 pattern (arrow) and one spermatozoa with a 10/12 pattern corresponding to an alternate or adjacent 1 segregation.
Discussion
Alternate and adjacent 1 segregations were the most common meiotic products found in this translocation as they accounted for 84.25% of the scored sperm. This incidence is in line with the frequencies found in most translocations [2, 8, 9]. As the normal chromosome and its translocated derivative cannot be distinguished by centromeric probes, the exact proportions of alternate and adjacent 1 could not be determined in this study. Sperm chromosome analysis of this translocation heterozygote yielded 61.1 and 26.3% alternate and adjacent ls, respectively [2]. To distinguish between alternate and adjacent 1 segregations, the application of a probe distal to the translocation breakpoint in one chromosome would be necessary.
Sperm resulting from adjacent 2 segregation were found at a frequency of 10.95% and were more common than 3:1s (4.42%). Compiled data from the literature show that most reciprocal translocations produce more adjacent 2 than 3:1s [2, 8]. This is in contrast with the frequencies of viable types of imbalances found at term. A study carried out on 161 reciprocal translocations showed an incidence of 66.5% for adjacent 1, 27.6% for 3:1, and 5.9% for adjacent 2 types of imbalances seen in liveborn [10]. While the number of translocations for which direct gamete data is available is in the order of 37, and thus much lower than the number of families from whom the incidences of imbalances at term have been drawn, the data are compatible with the existence of embryonic selection, sperm selection or both. The presence of embryonic selection in humans has been well established. Both the review of the European and USA collaborative prenatal diagnosis data have shown that the risk for unbalanced liveborn offspring is related to the extent of chromosome imbalance, with smaller imbalances having higher chances for survival [11, 12].
The incidence of 3:1 segregation in this patient was 4.42% and again was comparable to that found with chromosome analysis (5.6%). Sperm with one of the chromosomes involved in the translocation were more common than sperm with the complementary complement of three chromosomes. This finding could not be explained by a possible technical artifact caused by the different contrast of the orange and green signals in the blue DAPI background as both 10/- and -/12 cells were more frequent than their counterparts. Indeed, the same excess of sperm with the n/- pattern has been observed in three reciprocal translocations studied by FISH so far: t(l;l1) [9], t(6;l1) and t(2; 14) [8]. Rousseaux et al. [8] opted for citing a doubling of the hyperhaploidy rate as the 3:1 frequency and considering the excess of hypohaploidies a technical artifact. As more data become available on using FISH as an approach to meiotic segregation studies, it will become apparent whether technical differences, such as the type of probe labelling or differences in scoring criteria, may in fact impact on the results. Nuclei with disomy for chromosome 10 and 12 independent of the segregation of the translocation would be counted as 3:1 gametes, as the hybridization signals do not allow differentiation between normal and derivative 10 and 12. This would overestimate 3:1 segregation, although minimally, as disomy for chromosome 12 is 0.08% in control donors in this laboratory [15] and disomy for chromosome 10 has been reported to be 0.18% [13] and 0.32% [14] also in males with normal karyotypes.
Sperm with two copies of chromosome 10 and 12 (0.23%) are presumably diploid. Similar frequencies of diploid spermatozoa have been seen in a series of control donors (0.18%, unpublished results) and have also been reported by others (0.26%; ref. 16]. FISH analysis with dual color probes provides data on the number of diploid sperm as opposed to sperm chromosome analysis. Indeed, the hamster egg-human sperm fusion technique allows Polyspermic fertilization, thus jeopardizing the classification of dyspermic fertilization and the penetration by a diploid sperm. Theoretically, a spermatozoon with a 10/ 10/12/12 chromosome complement could also be the result of a 4:0 segregation. To truly separate diploids from 4:0s a triple color hybridization would be required. Rousseaux et al. [8] report incidences of diploidy of 0.21 % and of 4:0 of 0.12% and 0.44% in two heterozygotes carriers of the same t(6; 11) using a triple color hybridization. The high incidence of 4:0 segregants in this latter individual contrasts with data from the literature obtained with classical chromosome analysis. Only two gametes resulting from 4:0 segregations have been described: one in a double heterozygote [17] and one in a simple heterozygote [2].
FISH studies are performed on smears of washed ejaculate containing dead spermatozoa as well as living functional sperm whereas sperm chromosome analysis requires motile sperm capable of membrane fusion and pro-nuclear development. The data published by Spriggs et al. [9] on a t(1;11) heterozygote present a comparison of FISH and cytogenetic results. Their report and ours demonstrate the lack of sperm selection based on the chromosome content in the hamster egg-human sperm fusion system. Although these data cannot be extrapolated to human in vivo fertilization, they are consistent with the notion of chromosomally abnormal sperm not being selected against.
Although FISH methodology does not provide information on the full chromosome complement of the spermatozoon, it does allow for rapid screening of numerical aberrations in a representative number of cells. The study of gametes using FISH may provide answers to longstanding questions such as whether there are chromosome-specific rates of aneuploidy. FISH in sperm may become a tool in the large-scale analysis of chemicals or compounds that, through exposure, may affect nondisjunction in humans. Presently, FISH sperm nuclei is limited to the detection of numerical aberrations, however, if modified techniques are developed in the future, it could be applied to the assessment of chromosome breakage and thus its applications broadened.
References
Cans C, Cohen O, Mermet MA, Demongeot J, Jalbert P: Human reciprocal translocations: Is the unbalanced mode at birth predictable?. Hum Genet 1993;91:228–232.
Estop AM, Van Kirk V, Cieply K: Segregation analysis of four translocations, t(2;18), t(3;15), t(5;7), and t(10;12), by sperm chromosome studies and a review of the literature. Cytogen-et Cell Genet 1995;70:80–87.
Martin RH, Spriggs EL: Sperm chromosome complements in a man heterozygous for a reciprocal translocation 46,XY,t(9;13) (q21.1; q21.2) and a review of the literature. Clin Genet 1995;47:42–46.
Jalbert P, Sele B, Jalbert H: Reciprocal translocations: A way to predict the mode of unbalanced segregation by pachytene diagram drawing. A study of 151 human translocations. Hum Genet 1980;55:209–222.
Biggers JD, Whitten WK, Whittingham DG: The culture of mouse embryos in vitro; in Daniel JD (ed): Methods in mammalian embryology. San Francisco, Freeman, 1971, pp 68–116.
Wyrobek AJ, Alhorn T, Balhorn R, Stanker L, Pinkel D: Fluorescence in situ hybridization to Y chromosome in decondensed human sperm nuclei. Mol Reprod Dev 1990;27:200–208.
Kanji GK: 100 statistical tests. Newbury Park, Sage, 1993.
Rousseaux S, Chevret E, Monteil M, Cozzi J, Pelletier F, Devillard F, Lespinasse J, Sele B: Meiotic segregation in males heterozygote for reciprocal translocations: Analysis of sperm nuclei by two and three colour fluorescence in situ hybridization. Cytogenet Cell Genet 1995; 71:240–246.
Spriggs EL, Martin RH: Analysis of segregation in a human male reciprocal translocation carrier, t(1;11) (p36.3; q 13.1), by two-colour fluorescence in situ hybridization. Molec Reprod Devel 1994;38:247–250.
Jalbert P, Sele B: Factors predisposing to adjacent 2 and 3:1 disjunctions: Study of 161 human reciprocal translocations. J Med Genet 1979;16:467–478.
Daniel A, Boue A, Gallano P: Prospective risk in reciprocal translocation heterozygotes at amniocentesis as determined by potential chromosome imbalance sizes. Data of the European collaborative prenatal diagnosis centers. Prenat Diagn 1986;6:315–350.
Daniel A, Hook EB, Wulf G: Collaborative USA data on prenatal diagnosis for parental carriers of chromosome rearrangements: Risks of unbalanced progeny; in Daniel A (ed): The Cytogenetics of Mammalian Autosomal Rearrangements. New York, Liss, 1988, pp 73–162.
Bischoff FZ, Nguyen DD, Burt KJ, Shaffer LG: Estimates of aneuploidy using multicolor fluorescence in situ hybridization on human sperm. Cytogenet Cell Genet 1994;66:237–243.
Guttenbach M, Schakowski R, Schmid M: Incidence of chromosome 3, 7, 10, 11, 17 and X disomy in mature sperm nuclei as determined by nonradioactive in situ hybridization. Hum Genet 1994;93:7–12.
Estop AM, Cieply K, Aston C: Sperm chromosome aneuploidy studies by FISH in men with normal karyotypes and translocation carriers (abstract). American Cytogenetics Conference, Seven Springs 1996. Cytogenet Cell Genet, in press.
Williams BJ, Ballenger CA, Malter HE, Bishop F, Tucker M, Zwingman TA, Hassold TJ: Nondisjunction in human sperm: results of fluorescence in situ hybridization studies using two and three probes. Hum Molec Genet 1993;2: 1929–1936.
Burns JP, Koduru PRK, Alonso ML, Chaganti RSK: Analysis of meiotic segregation in a man heterozygous for two reciprocal translocations using the hamster in vitro penetration system. Am J Hum Genet 1986;38:954–964.
Acknowledgment
We are grateful to Cheryl Cummings for assistance in the preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Estop, A.M., Cieply, K.M. & Aston, C.E. The Meiotic Segregation Pattern of a Reciprocal Translocation t(10;12)(q26.1; p13.3) by Fluorescence in situ Hybridization Sperm Analysis. Eur J Hum Genet 5, 78–82 (1997). https://doi.org/10.1007/BF03405881
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03405881