Abstract
Copy number variation (CNV) remains poorly defined in many populations, including Mexican Americans. We report the discovery and genetic confirmation of copy number variable regions (CNVRs) in subjects of the San Antonio Family Heart and the San Antonio Family Diabetes Gallbladder Studies, both comprised of multigenerational pedigrees of Mexican American descent. In a discovery group of 1677 participants genotyped using Illumina Infinium Beadchips, we identified 2937 unique CNVRs, some with observation frequencies as low as 0.002, using a process that integrates pedigree information with CNV calls made by PennCNV and/or QuantiSNP. Quantitative copy number values had statistically significant (P≤1.792e-5) heritability estimates ranging from 0.139 to 0.863 for 2776 CNVRs. Additionally, 920 CNVRs showed evidence of linkage to their genomic location, providing strong genetic confirmation. Linked CNVRs were enriched in a set of independently identified CNVRs from a second group of 380 samples, confirming that these CNVRs can be used as predefined CNVRs of high confidence. Interestingly, we identified 765 putatively novel variants that do not overlap with the Database of Genomic Variants. This study is the first to use linkage and heritability in multigenerational pedigrees as a confirmation approach for the discovery of CNVRs, and the largest study to date investigating copy number variation on a genome-wide scale in individuals of Mexican American descent. These results provide insight to the structural variation present in Mexican Americans and show the strength of multigenerational pedigrees to elucidate structural variation in the human genome.
Similar content being viewed by others
Introduction
Copy number variants (CNVs), gains or losses of DNA sequence larger than 1 kb, were first recognized as widespread in the human genome in 2004.1 Since this initial discovery multiple studies have further characterized copy number variation in the human genome.2, 3, 4 Recent reports suggest that CNVs have a role in multiple complex disorders, such as schizophrenia,5 autism,6 autoimmune disorders,7 and diabetes syndromes.7, 8
However, despite this progress, copy number variation remains poorly defined in many populations. Understanding genetic variation in human populations besides Caucasians may reveal important biological insights not observable in the Caucasian population and is important for extending the benefits of understanding genetic risk to these underrepresented populations.
Despite methodological advancements for identifying, genotyping, and characterizing CNVs,9 there is currently no comprehensive cost-effective method that has reached universal adoption. Several studies, including HapMap3,4 have recently used the Illumina Infinium technology to characterize CNVs.
Given the current limitations of CNV analysis, most studies have taken the approach of limiting type I error by either requiring CNVs to be identified by more than one algorithm, or by limiting the size or number of probes identifying a CNV so that an acceptable portion are confirmed by orthogonal approaches. However, investigating copy number variation in multigenerational pedigrees allows for additional quality control metrics such as observation of transmission, and linkage with adjacent markers to confirm the genomic location of CNVs. Additionally, some rare CNVs will be present in the founders and may be inherited by younger generations in the pedigree, thus allowing for higher confidence in rare CNV identification. Despite these strengths, there are currently no large-scale studies reporting CNVs identified in multigenerational cohorts that use this information as a form of quality control.
In this study, we investigate copy number variation in 2057 participants of the San Antonio Family Heart Study (SAFHS) and San Antonio Family Diabetes/Gallbladder Study (SAFDGS), both comprised of multigenerational pedigrees of Mexican American descent. We present the identification, genotyping, and confirmation of copy number variable regions (CNVRs) using heritability and linkage. We report their genomic distribution, potential disease relevance, and discovery of novel variants. Most importantly, this study provides novel insight into the structural variation specific to Mexican Americans.
Materials and methods
Study design
Participants in this study are members of extended, multigenerational families of Mexican American descent who have taken part in the SAFHS or the SAFDGS. Study-related clinical exams were conducted in San Antonio, Texas. SAFHS is a family study where the subjects were not ascertained on disease status. SAFDGS probands were ascertained on type 2 diabetes status. The current study was approved by the Institutional Review Board at the University of Texas Health Science Center San Antonio, and informed consent was obtained from all participants. Both cohorts have previously been described in detail.10, 11
Study group 1
DNA isolated from primary blood mononuclear cells for 1677 participants was previously genotyped using four versions of the Illumina (San Diego, CA, USA) Infinium Beadchips: 767 participants were genotyped on the 1M duo beadchip, 327 individuals were genotyped on the 1M beadchip, and 583 individuals were genotyped on both the 510 and 550 beadchips. The SNP markers on the 510 and 550 beadchips are unique and together represent the content on the 1M beadchip.
Study group 2
Participants of this group are members of the SAFDGS for which DNA from lymphoblastoid cell lines were genotyped on the Illumina 660W beadchip. Data from a total of 380 participants were available for analyses.
CNVR identification
Approach
We first applied a ‘wide’ method of identification of CNVs to these data by applying CNV-calling algorithms to identify CNVs in individual samples. As these methods are known to have high rates of type I and type II errors, we also reassigned copy number values to individual participants based on comparison of the samples to each other, described in the section ‘CNVR genotyping’ referred to here as a ‘deep’ method.
To minimize type II errors that may be present using one algorithm, we chose to employ two algorithms, PennCNV12 and QuantiSNP.13 Standard quality control measures implemented within PennCNV were utilized for sample exclusion. The Log ratio SD, b-allele ratio drift, and genomic waviness were set at maximums of 0.3, 0.01, and 0.05, respectively. QuantiSNP calls were limited to CNV calls with a Maximum Bayesian Factor ≥10.
Recent CNV studies have taken the approach of reporting only those CNVs that are similarly identified by at least two algorithms in the same individual.14 Given that our downstream analyses could be used to identify type I errors, we chose to take a more liberal approach to CNV inclusion so as to identify as much variation as possible. As pedigree data gives us additional information by which we could validate CNVs, we took the approach of including CNVs even if they are only identified by one algorithm.
Using the individual CNV calls, CNVRs, which are regions of overlap of CNVs, were identified. To reduce the rate of type I errors we limited our downstream analysis to regions harboring a CNV call in at least two individuals of the same pedigree on the same beadchip. To determine breakpoints for this set of CNVRs, we expanded the CNV breakpoints to the largest overlapping region identified in each pedigree. This set of CNVRs we have termed Pedigree CNVRs. To further summarize the observed CNV calls, we consolidated CNVRs across all pedigrees by using the most common breakpoints observed for overlapping Pedigree CNVRs. For those consolidated CNVRs that were observed to be overlapping on multiple platforms, we used the breakpoints identified by the higher density beadchips. Priority was given to breakpoints identified using PennCNV, when the CNV was identified by both algorithms. Each CNVR was then manually inspected to ensure the individual CNV calls indicated the identified CNVR breakpoints, which resulted in removing five CNVRs and redefining the breakpoints for CNVR887. All other CNVRs remained unchanged.
CNVR genotyping
Considering each beadchip independently, for each final CNVR we used CNVtools15 to identify the first principal component of log R ratios of markers falling within the CNVR breakpoints. Using this value, we used CNVtools to cluster individuals into groups harboring the same copy number genotype. To improve our power for downstream analyses, we combined the Log R ratio data from the 1M duo, 1 M, 550, and 510 beadchips into a single matrix and performed the same procedures using CNVtools.
CNVR characterization
PennCNV calls and histograms of the first principal component were used to categorize each CNVR as a deletion, duplication, complex, overlapping, or unknown. CNVRs were considered overlapping if there were two clear variants of different lengths, either duplication or deletion, which were both overlapping and present in multiple individuals. Complex regions were labeled based on their location in either centromeric, telomeric, or immunoglobulin regions. Unknown regions were those that we were unable to clearly classify based on the available data, but may fit into any of the other four categories. Tables from the UCSC genome browser summarizing OMIM genes, RefSeq genes, segmental duplications, microRNAs, and disease association SNPs from the National Human Genome Research Institute were downloaded on 1 March 2011 and used as reference datasets. A table from the UCSC genome browser summarizing the Database of Genomic Variants (DGVs) was downloaded on 4 February 2012 and used as a reference dataset. Copy Number Polymorphisms were downloaded from HapMap3 on 23 March 2011 for comparison of lengths.
Heritability and linkage analysis
For each CNVR, the first principal component value identified by CNVtools was rank normalized. Subsequently, heritability was calculated and each CNVR was tested for linkage to its own genomic location. Both tests were conducted using a variance components approach using the statistical models implemented within the software package SOLAR.16, 17 Heritability and linkage was also calculated for binned copy number values by treating the values as a categorical trait, either harboring or not harboring a deletion or duplication.
Statistical analyses of results
CNVR lengths and minor allele frequencies were log transformed to provide a normal distribution. Correlation of lengths between this study and HapMap3 was tested using linear regression. Correlations between the observed minor allele frequency and heritability or linkage LOD scores were tested using linear regression. The difference in lengths between deletions and duplications was tested using a one-sided t-test. The difference in ratios of deletions and duplications which overlap genes was tested using a χ2 test. The correlation between length and observed minor allele frequency of deletions was tested using a linear model. The difference in size between novel and known CNVRs was tested using a two sided t-test. CNVRs that were linked to their genomic location in group 1were tested for enrichment in group 2 using a χ2 test.
Results
Study group 1
We identified 2937 unique CNVRs, representing 120 959 and 75 932 autosomal CNV calls by either PennCNV or QuantiSNP, respectively, detected across all individuals genotyped on any SNP microarray type. In all, 1201 CNVRs are pedigree specific, 399 of which have not been previously reported in the DGVs and potentially represent private variants enriched within the pedigrees through transmission. Summary information for CNV calls is presented in Table 1.
When applying the Gaussian mixture model implemented within CNVtools,15 we were able to confidently fit 186 CNVRs into defined classes. When coded as a dichotomous trait, 169 (90.9%) of these CNVRs had a statistically significant (P≤2.7e-4) heritability of 1.00, as would be expected for a correctly genotyped copy number variant. Ten additional CNVRs had statistically significant (P≤2.7e-4) heritabilities ranging from 0.723 to 0.977. In all, 151 (81.2%) CNVRs showed evidence of linkage to their genomic location (P≤0.05), 131 (70.4%) of which are linked after Bonferroni correction for the number of CNVRs investigated (P≤2.7e-4).
Given the high ratio of observed posterior probability errors when fitting these measurements into defined bins, we chose to work with the first principal component as a measurement of copy number as it was available for all 2937 CNVRs, an approach that has been used previously.18 For 2776 (94.5%) CNVRs the first principal component had statistically significant (P≤1.702e-5) heritabilities ranging from 0.139 to 0.863. For 920 (31.3%) CNVRs the first principal component showed evidence of linkage to its genomic location (P≤0.05). Of 727 novel CNVRs, 670 (92.2%) are significantly heritable, and 146 (20.1%) show evidence of linkage (P≤0.05), providing very strong evidence of the validity of these novel variants. Linkage and heritability information for different classes of variants is presented in Table 2.
We hypothesized that rare variants may not be sufficiently measured by the first principal component value for significant linkage to be observed. We investigated the relationship between the observation frequency of 186 CNVRs that were binned into defined classes and their respective heritability and linkage LOD values using the first principal component. These CNVRs had observation frequencies ranging from 0.002 to 0.477. Observation frequency was positively correlated with heritability (P≤2e-16), as shown in Figure 1. Observation frequency was also associated with linkage LOD values (P≤2e-16), supporting our hypothesis that common variants were more likely to be linked in our analysis. On the basis of this observation and the observation that dispersed duplications may insert elsewhere in the genome, we conclude that lack of linkage to their genomic location does not indicate that a CNVR is the result of a type 1 error. Rather we consider those that have statistically significant heritability estimates to be confirmed, and those that are linked to be of the highest confidence, having evidence of their genomic location. The location, class, heritability estimates, and linkage LOD values for all CNVRs are presented in the Supplementary Materials.
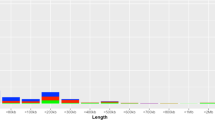
The lengths of the CNVRs identified in this study correlated significantly (P≤2.2e-16) with lengths of overlapping CNVRs from HapMap3 (392 CNVRs).4 Visual inspection, presented in Figure 2, indicates a generally good agreement of lengths between studies. When considering those CNVRs that show evidence of linkage (P<0.05), the average length of deletions and duplications are 21.8 and 45.5 kb, respectively (P=4.62E-5), as shown in Figure 3. Twenty-nine of 178 (16.3%) duplications encompass at least 1 gene compared with 25 of 505 (5.0%) deletions (P=1.42E-6), but this difference is not statistically significant when corrected for the length of the CNVRs. Interestingly, within 186 CNVRs, which were binned into defined states, large deletions had lower observation frequencies than smaller deletions (P=1.2E-8).
Size distribution of deletions and duplications linked to their genomic location. The size distribution of duplications is presented in transparent white and is superimposed over the size distribution of deletions, which is presented in dark gray. There is a statistically significant difference in size distribution likely representing an improved ability to detect small deletions.
Four-hundred and nine CNVRs identified in this study are within 50 kb of a disease associated SNP from the NHGRI GWAS catalog. Among these, was a CNVR harboring two overlapping deletions, a common ∼1-kb deletion within the ACACA gene and a ∼1.44-Mb deletion of 16 genes, including HNF1B, which is responsible for Renal Cyst and Diabetes Syndrome, also referred to as Maturity Onset Diabetes of the Young 5 (MODY5[MIM 137920]). This is a recurrent deletion that has been associated with multiple phenotypes including MODY5 and psychiatric disorders.19, 20, 21 The deletion was apparent in three individuals, a woman and her two daughters. We hypothesized that these individuals had MODY5. Retrospective investigation of clinical data showed that the mother and one daughter were diagnosed with diabetes at ages 17 and 22.4 years, respectively. One daughter remained diabetes free at her last visit at age 31, indicating incomplete penetrance.
We discovered 727 putatively novel CNVRs that do not overlap with variants reported in the DGVs.1 Of these, 328 were detected in multiple pedigrees. These CNVRs may have been missed in previous studies or may be unique to the Mexican American population in this study. Considering CNVRs, which showed evidence of linkage to their genomic location (P≤0.05), novel CNVRs identified in this study were smaller on average than previously known CNVRs (P=0.0004), as shown in Figure 4.
Of 146 novel and linked CNVRs, 21 are within 50 kb of a disease associated SNP from the NHGRI. Of these 21, 10 overlap with gene exons, providing novel testable hypotheses, which are summarized in the Supplementary Materials. One of these 10 is a ∼5.2-kb deletion of the IL2 gene. This CNVR is significantly heritable (P=6.35E-12), linked (P=0.045), and is flanked by SNPs associated with immune-related functions.22, 23 A second of these 10 is a 370-bp CNVR within the first exon of the UGT1A7 gene. This CNVR is highly linked to its own genomic location (P=5.44E-60), and ∼6.6 kb from rs2602381, which was previously associated with attention-deficit hyperactivity disorder.24
Study group 2
We identified 2555 CNVRs representing 106 219 and 116 505 autosomal CNV calls using PennCNV and QuantiSNP, respectively. Of 72 putatively novel CNVRs discovered in this group, 34 were also discovered in group 1. Despite being identified using separate platforms, CNVRs in group 2 overlap 745 CNVRs identified in Study group 1, of which 420 (56.4%) showed evidence of linkage (P≤0.05) in group 1. This enrichment is statistically significant (P≤1.0e-10), showing that those which were linked in the first study were more likely to be observed in a second study group, as well as validating linkage as a useful confirmation approach.
Discussion
Copy number variation makes up a significant portion of genetic variation in humans. The current limitations regarding CNVs are largely due to a lack of an affordable comprehensive identification and genotyping strategy, although methods have been proposed to address this issue.9, 25 We have applied a strategy that takes advantage of the benefit of pedigree information to identify, confirm, and localize CNVs in the largely understudied Mexican American population. In an effort to limit type 1 errors, previous reports have limited their analysis to CNVs identified using at least two algorithms in the same individual. However, in this study we were able to rely on pedigree information as a form of quality control, so we did not restrict CNVRs based on this convention. Of 431 CNVRs which we have confirmed beyond doubt using linkage analysis, 144 CNVRs were identified by PennCNV only and 21 CNVRs were identified by QuantiSNP only in at least two individuals in the same pedigree, indicating that restricting CNVRs based on algorithm overlap can be overly conservative.
Similar to previous reports,7, 18 we observed poor cluster separation for many regions and were unable to confidently bin individuals into defined classes. However, we are able to show that for ∼95% of the CNVRs identified here, representative values for these regions had statistically significant heritability estimates. Additionally, 920 CNVRs showed evidence of linkage to their genomic location, providing exceptionally strong genetic confirmation. This nicely highlights the continued difficulty of genotyping CNVs, and supports the use of representative values in the absence of high-confidence binning.
Through linkage we obtained evidence that 178 duplications have inserted near their genomic location. This does not mean that these are tandem duplications, because a dispersed duplication could potentially be close enough to its original genomic location to be linked. The extent to which dispersed duplications are responsible for associations on other chromosomes is currently unknown. Future studies aimed at using linkage analysis in multigenerational pedigrees may help to identify the insertion locations of common duplications.
The observed difference in size between duplications and deletions in this study could be the result of a methodological bias toward detecting large CNVs. Similarly, the correlation between deletion size and frequency may indicate an increased ability to detect large deletions. Alternatively, these observations could indicate that large deletions are under stronger selective pressure than duplications or small deletions.
In summary, we have identified and genotyped CNVRs that are polymorphic in Mexican Americans from San Antonio, Texas. The majority of CNVRs identified have been previously reported, indicating Mexican Americans share much of their genetic diversity with other populations. However, about 25% of copy number variation in this population may be specific to this ethnic group and has not been previously characterized due to the limited number of studies in Mexican American populations.
Importantly, we show that CNVRs that were confirmed using linkage analysis in this study are likely to be identified again in a separate study, and therefore can be used as predefined CNVRs of high confidence in future studies investigating CNVs in Mexican Americans. Our application of heritability and linkage analysis to confirm CNVR genotype measurements shows the promise of using multigenerational pedigrees to improve the power and accuracy with which we can characterize structural variation in the human genome, and should be considered orthogonal to other quality metrics for CNV calling. We suggest that future studies investigating copy number variation in multigenerational pedigrees should incorporate similar approaches to for CNVR confirmation.
References
Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Redon R, Ishikawa S, Fitch KR et al: Global variation in copy number in the human genome. Nature 2006; 444: 444–454.
Conrad DF, Pinto D, Redon R et al: Origins and functional impact of copy number variation in the human genome. Nature 2010; 464: 704–712.
Altshuler DM, Gibbs RA, Peltonen L et al: Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52–58.
Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M : Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet 2008; 40: 880–885.
Sebat J, Lakshmi B, Malhotra D et al: Strong association of de novo copy number mutations with autism. Science 2007; 316: 445–449.
Craddock N, Hurles ME, Cardin N et al: Genome-wide association study of CNVs in 16 000 cases of eight common diseases and 3000 shared controls. Nature 2010; 464: 713–720.
Jarick I, Vogel CI, Scherag S et al: Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum Mol Genet 2011; 20: 840–852.
Alkan C, Coe BP, Eichler EE : Genome structural variation discovery and genotyping. Nat Rev Genet 2011; 12: 363–376.
Hunt KJ, Lehman DM, Arya R et al: Genome-wide linkage analyses of type 2 diabetes in Mexican Americans: the San Antonio Family Diabetes/Gallbladder Study. Diabetes 2005; 54: 2655–2662.
Mitchell BD, Kammerer CM, Blangero J et al: Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation 1996; 94: 2159–2170.
Wang K, Li M, Hadley D et al: PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007; 17: 1665–1674.
Colella S, Yau C, Taylor JM et al: QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res 2007; 35: 2013–2025.
Wineinger NE, Pajewski NM, Kennedy RE et al: Characterization of autosomal copy-number variation in African Americans: the HyperGEN Study. Eur J Hum Genet 2011.
Barnes C, Plagnol V, Fitzgerald T et al: A robust statistical method for case-control association testing with copy number variation. Nat Genet 2008; 40: 1245–1252.
Boerwinkle E, Chakraborty R, Sing CF : The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet 1986; 50: 181–194.
Almasy L, Blangero J : Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62: 1198–1211.
Surakka I, Kristiansson K, Anttila V et al: Founder population-specific HapMap panel increases power in GWA studies through improved imputation accuracy and CNV tagging. Genome Res 2010; 20: 1344–1351.
Nagamani SC, Erez A, Shen J et al: Clinical spectrum associated with recurrent genomic rearrangements in chromosome 17q12. Eur J Hum Genet 2010; 18: 278–284.
Mefford HC, Clauin S, Sharp AJ et al: Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet 2007; 81: 1057–1069.
Moreno-De-Luca D, Mulle JG, Kaminsky EB et al: Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet 2010; 87: 618–630.
Plagnol V, Howson JM, Smyth DJ et al: Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 2011; 7: e1002216.
Ramasamy A, Curjuric I, Coin LJ et al: A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol 2011; 128: 996–1005.
Mick E, Todorov A, Smalley S et al: Family-based genome-wide association scan of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 2010; 49: e893.
Park H, Kim JI, Ju YS et al: Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet 2010; 42: 400–405.
Acknowledgements
This study was supported in part by grants from the National Institutes of Health (DK47482, DK70746, DK053889, HLO45222, RR013556, MH059490) and the Department of Defense (DOD PC081025). We thank the participants of the SAFHS and SAFDGS for their generous cooperation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Blackburn, A., Göring, H., Dean, A. et al. Utilizing extended pedigree information for discovery and confirmation of copy number variable regions among Mexican Americans. Eur J Hum Genet 21, 404–409 (2013). https://doi.org/10.1038/ejhg.2012.188
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.188
Keywords
This article is cited by
-
Characterization of Large Copy Number Variation in Mexican Type 2 Diabetes subjects
Scientific Reports (2017)
-
Imputation in families using a heuristic phasing approach
BMC Proceedings (2014)