Abstract
CYP2A6 metabolically inactivates nicotine. Faster CYP2A6 activity is associated with heavier smoking and higher lung cancer risk. The CYP2A6 gene is polymorphic, including functional structural variants (SV) such as gene deletions (CYP2A6*4), duplications (CYP2A6*1 × 2), and hybrids with the CYP2A7 pseudogene (CYP2A6*12, CYP2A6*34). SVs are challenging to genotype due to their complex genetic architecture. Our aims were to develop a reliable protocol for SV genotyping, functionally phenotype known and novel SVs, and investigate the feasibility of CYP2A6 SV imputation from SNP array data in two ancestry populations. European- (EUR; n = 935) and African- (AFR; n = 964) ancestry individuals from smoking cessation trials were genotyped for SNPs using an Illumina array and for CYP2A6 SVs using Taqman copy number (CN) assays. SV-specific PCR amplification and Sanger sequencing was used to characterize a novel SV. Individuals with SVs were phenotyped using the nicotine metabolite ratio, a biomarker of CYP2A6 activity. SV diplotype and SNP array data were integrated and phased to generate ancestry-specific SV reference panels. Leave-one-out cross-validation was used to investigate the feasibility of CYP2A6 SV imputation. A minimal protocol requiring three Taqman CN assays for CYP2A6 SV genotyping was developed and known SV associations with activity were replicated. The first domain swap CYP2A6-CYP2A7 hybrid SV, CYP2A6*53, was identified, sequenced, and associated with lower CYP2A6 activity. In both EURs and AFRs, most SV alleles were identified using imputation (>70% and >60%, respectively); importantly, false positive rates were <1%. These results confirm that CYP2A6 SV imputation can identify most SV alleles, including a novel SV.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Full exon and breakpoint DNA sequences for the CYP2A6*53 SV are available in Supplementary Fig. 1.
References
American Lung Association. Lung Cancer Fact Sheet | American Lung Association [Internet]. Chicago, IL: American Lung Association; 2022 [updated 2022 November 17; cited 2023 February 22]. Available from: https://www.lung.org/lung-health-diseases/lung-disease-lookup/lung-cancer/resource-library/lung-cancer-fact-sheet.
McDonagh EM, Wassenaar C, David SP, Tyndale RF, Altman RB, Whirl-Carrillo M, et al. PharmGKB summary: very important pharmacogene information for cytochrome P-450, family 2, subfamily A, polypeptide 6. Pharmacogenet Genomics. 2012;22:695–708.
Benowitz NL. Clinical pharmacology of nicotine: implications for understanding, preventing, and treating tobacco addiction. Clin Pharm Ther. 2008;83:531–41.
Dempsey D, Tutka P, Jacob P, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharm Ther. 2004;76:64–72.
Lerman C, Tyndale R, Patterson F, Wileyto EP, Shields PG, Pinto A, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharm Ther. 2006;79:600–8.
Benowitz NL, Pomerleau OF, Pomerleau CS, Jacob P. Nicotine metabolite ratio as a predictor of cigarette consumption. Nicotine Tob Res. 2003;5:621–4.
Wassenaar CA, Ye Y, Cai Q, Aldrich MC, Knight J, Spitz MR, et al. CYP2A6 reduced activity gene variants confer reduction in lung cancer risk in African American smokers-findings from two independent populations. Carcinogenesis. 2015;36:99–103.
Byun J, Han Y, Li Y, Xia J, Long E, Choi J, et al. Cross-ancestry genome-wide meta-analysis of 61,047 cases and 947,237 controls identifies new susceptibility loci contributing to lung cancer. Nat Genet. 2022;54:1167–77.
Liu T, David SP, Tyndale RF, Wang H, Zhou Q, Ding P, et al. Associations of CYP2A6 genotype with smoking behaviors in southern China. Addiction. 2011;106:985–94.
Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103:1342–6.
El-Boraie A, Taghavi T, Chenoweth MJ, Fukunaga K, Mushiroda T, Kubo M, et al. Evaluation of a weighted genetic risk score for the prediction of biomarkers of CYP2A6 activity. Addict Biol. 2020;25:e12741.
El-Boraie A, Chenoweth MJ, Pouget JG, Benowitz NL, Fukunaga K, Mushiroda T, et al. Transferability of ancestry-specific and cross-ancestry CYP2A6 activity genetic risk scores in African and European populations. Clin Pharmacol Ther. 2021;110:975–85.
Nunoya K, Yokoi T, Kimura K, Inoue K, Kodama T, Funayama M, et al. A new deleted allele in the human cytochrome P450 2A6 (CYP2A6) gene found in individuals showing poor metabolic capacity to coumarin and (+)-cis-3,5-dimethyl-2-(3-pyridyl)thiazolidin-4-one hydrochloride (SM-12502). Pharmacogenetics. 1998;8:239–49.
Oscarson M, McLellan RA, Asp V, Ledesma M, Bernal Ruiz ML, Sinues B, et al. Characterization of a novel CYP2A7/CYP2A6 hybrid allele (CYP2A6*12) that causes reduced CYP2A6 activity. Hum Mutat. 2002;20:275–83.
Rao Y, Hoffmann E, Zia M, Bodin L, Zeman M, Sellers EM, et al. Duplications and defects in the CYP2A6 gene: identification, genotyping, and in vivo effects on smoking. Mol Pharm. 2000;58:747–55.
di Iulio J, Fayet A, Arab-Alameddine M, Rotger M, Lubomirov R, Cavassini M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genomics. 2009;19:300–9.
Pharmacogene Variation Consortium. CYP2A6 [Internet]. Kansas City, MO: Pharmacogene Variation Consortium; 2022 [updated 2022 October 11; cited 2023 February 22]. Available from: https://www.pharmvar.org/gene/CYP2A6.
Chen LS, Hartz SM, Baker TB, Ma Y, Saccone L, Bierut N, et al. Use of polygenic risk scores of nicotine metabolism in predicting smoking behaviors. Pharmacogenomics. 2018;19:1383–94.
Wassenaar CA, Zhou Q, Tyndale RF. CYP2A6 genotyping methods and strategies using real-time and end point PCR platforms. Pharmacogenomics. 2016;17:147–62.
Langlois AWR, El-Boraie A, Fukunaga K, Mushiroda T, Kubo M, Lerman C, et al. Accuracy and applications of sequencing and genotyping approaches for CYP2A6 and homologous genes. Pharmacogenet Genomics. 2022;32:159–72.
Cox LS, Faseru B, Mayo MS, Krebill R, Snow TS, Bronars CA, et al. Design, baseline characteristics, and retention of African American light smokers into a randomized trial involving biological data. Trials. 2011;12:22.
Nollen NL, Cox LS, Yu Q, Ellerbeck EF, Scheuermann TS, Benowitz NL, et al. A clinical trial to examine disparities in quitting between African-American and White adult smokers: design, accrual, and baseline characteristics. Contemp Clin Trials. 2016;47:12–21.
Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–83.
Leffler EM, Band G, Busby GBJ, Kivinen K, Le QS, Clarke GM, et al. Resistance to malaria through structural variation of red blood cell invasion receptors. Science. 2017;356:eaam6393.
Häkkinen K, Kiiski JI, Lähteenvuo M, Jukuri T, Suokas K, Niemi-Pynttäri J, et al. Implementation of CYP2D6 copy-number imputation panel and frequency of key pharmacogenetic variants in Finnish individuals with a psychotic disorder. Pharmacogenomics J. 2022;22:166–72.
Boettger LM, Salem RM, Handsaker RE, Peloso GM, Kathiresan S, Hirschhorn JN, et al. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat Genet. 2016;48:359–66.
Lerman C, Schnoll RA, Hawk LW, Cinciripini P, George TP, Wileyto EP, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respir Med. 2015;3:131–8.
Cox LS, Nollen NL, Mayo MS, Choi WS, Faseru B, Benowitz NL, et al. Bupropion for smoking cessation in African American light smokers: a randomized controlled trial. J Natl Cancer Inst. 2012;104:290–8.
Chenoweth MJ, Ware JJ, Zhu AZX, Cole CB, Cox LS, Nollen N, et al. Genome-wide association study of a nicotine metabolism biomarker in African American smokers: impact of chromosome 19 genetic influences. Addiction. 2018;113:509–23.
Johansson LF, van Dijk F, de Boer EN, van Dijk-Bos KK, Jongbloed JD, van der Hout AH, et al. CoNVaDING: single exon variation detection in targeted NGS data. Hum Mutat. 2016;37:457–64.
Gaedigk A, Casey ST, Whirl-Carrillo M, Miller NA, Klein TE. Pharmacogene Variation Consortium: a global resource and repository for pharmacogene variation. Clin Pharm Ther. 2021;110:542–5.
Zhao S, Jing W, Samuels DC, Sheng Q, Shyr Y, Guo Y. Strategies for processing and quality control of Illumina genotyping arrays. Brief Bioinform. 2018;19:765–75.
Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–97.
Sicko RJ, Romitti PA, Browne ML, Brody LC, Stevens CF, Mills JL, et al. Rare variants in RPPH1 real-time quantitative PCR control assay binding sites result in incorrect copy number calls. J Mol Diagn. 2022;24:33–40.
Nakano M, Fukushima Y, Yokota S, Fukami T, Takamiya M, Aoki Y, et al. CYP2A7 pseudogene transcript affects CYP2A6 expression in human liver by acting as a decoy for miR-126. Drug Metab Dispos. 2015;43:703–12.
Nofziger C, Turner AJ, Sangkuhl K, Whirl-Carrillo M, Agúndez JAG, Black JL, et al. PharmVar GeneFocus: CYP2D6. Clin Pharm Ther. 2020;107:154–70.
Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–12.
Reid BM, Permuth JB, Chen YA, Fridley BL, Iversen ES, Chen Z, et al. Genome-wide analysis of common copy number variation and epithelial ovarian cancer risk. Cancer Epidemiol Biomark Prev. 2019;28:1117–26.
Walker LC, Marquart L, Pearson JF, Wiggins GA, O’Mara TA, Parsons MT, et al. Evaluation of copy-number variants as modifiers of breast and ovarian cancer risk for BRCA1 pathogenic variant carriers. Eur J Hum Genet. 2017;25:432–8.
Haberl M, Anwald B, Klein K, Weil R, Fuss C, Gepdiremen A, et al. Three haplotypes associated with CYP2A6 phenotypes in Caucasians. Pharmacogenet Genomics. 2005;15:609–24.
Pang C, Liu JH, Xu YS, Chen C, Dai PG. The allele frequency of CYP2A6*4 in four ethnic groups of China. Exp Mol Pathol. 2015;98:546–8.
Claw KG, Beans JA, Lee SB, Avey JP, Stapleton PA, Scherer SE, et al. Pharmacogenomics of nicotine metabolism: novel CYP2A6 and CYP2B6 genetic variation patterns in Alaska Native and American Indian populations. Nicotine Tob Res. 2020;22:910–8.
Yadav VK, Katiyar T, Ruwali M, Yadav S, Singh S, Hadi R, et al. Polymorphism in cytochrome P4502A6 reduces the risk to head and neck cancer and modifies the treatment outcome. Environ Mol Mutagen. 2021;62:502–11.
Takeshita H, Hieda Y, Fujihara J, Xue Y, Nakagami N, Takayama K, et al. CYP2A6 polymorphism reveals differences in Japan and the existence of a specific variant in Ovambo and Turk populations. Hum Biol. 2006;78:235–42.
Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74.
Acknowledgements
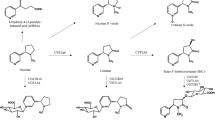
We acknowledge the contributions of Dr. Caryn Lerman in the design and administration of the Pharmacogenetics of Nicotine Addiction Treatment 2 clinical trial (NCT01314001), of Dr. Michiaki Kubo in contributing to the deep exon sequencing and initial Taqman CNV assay genotyping of participants, and of Dr. Andrea Gaedigk and the CYP2A6 Pharmvar expert panel in naming the novel structural variant, CYP2A6*53, and inspiring the CYP2A6 structural variant schematics in Fig. 1. This research was undertaken, in part, thanks to funding from the Canada Research Chairs program (Tyndale, the Canada Research Chair in Pharmacogenomics), Canadian Institutes of Health Research (Tyndale, FDN-154294; Tyndale, Knight, and Chenoweth, PJY-159710); National Institute on Drug Abuse (Tyndale, PGRN U01-DA20830), and National Institutes of Cancer (Cox, NCI CA091912) as well as the Centre for Addiction and Mental Health and the CAMH Foundation. JSA funded in part by P20GM130414, a NIH funded Center of Biomedical Research Excellence (COBRE).
Author information
Authors and Affiliations
Contributions
AL, JP, JK, MC, and RT designed the research; AE, LC, JA, KF, and TM provided resources, AL performed the research; AL analyzed the data; AL and RT wrote the manuscript; all authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
Some of the authors declared financial and non-financial relationships and activities, and conflicts of interest regarding this manuscript as indicated in the Supplementary Materials.
Ethical approval
Ethical approval for these analyses was obtained from the University of Toronto (PNAT2 REB number: 25510; KIS3 REB number: 38361) and at all original participating sites of the trials.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Langlois, A.W.R., El-Boraie, A., Pouget, J.G. et al. Genotyping, characterization, and imputation of known and novel CYP2A6 structural variants using SNP array data. J Hum Genet 68, 533–541 (2023). https://doi.org/10.1038/s10038-023-01148-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01148-y
This article is cited by
-
Associating CYP2A6 structural variants with ovarian and lung cancer risk in the UK Biobank: replication and extension
European Journal of Human Genetics (2024)