Abstract
Non-small cell lung cancer (NSCLC) has high rates of morbidity and mortality. E3 ubiquitin ligase usually has antitumor effects. This study evaluated the mechanism of E3 ligase FBXW7 (F-box and WD repeat domain containing 7) in the radiosensitivity of NSCLC. NCI-H1299 and NCI-H1299R cells were irradiated by 0, 2, 4, and 6 Gy doses of X-ray, respectively. In addition to the measurement of cell proliferation, apoptosis, and γ-H2AX, FBXW7 expression was measured and the interaction between FBXW7 and SOX9 (SRY-box transcription factor 9) was evaluated. Ubiquitination level and protein stability of SOX9 were examined after FBXW7 overexpression. The binding relationship between SOX9 and CDKN1A (cyclin-dependent kinase inhibitor 1A) was verified. Xenograft tumor model was established to evaluate the effect of FBXW7 on radiosensitivity in vivo. FBXW7 was under-expressed in radioresistant cells. Overexpression of FBXW7 repressed NCI-H1299 and NCI-H1299R cell proliferation and colony formation and increased γ-H2AX-positive foci. Overexpression of FBXW7 increased the ubiquitination level and reduced the protein stability of SOX9. SOX9 bound to the CDKN1A promoter to inhibit CDKN1A expression. FBXW7 inhibited tumorigenesis and apoptosis and enhanced radiosensitivity of NSCLC cells in vivo via the SOX9/CDKN1A axis. Overall, FBXW7 inhibited SOX9 expression by promoting SOX9 ubiquitination and proteasome degradation, suppressing the binding of SOX9 to CDKN1A, and upregulating CDKN1A, thereby improving the radiosensitivity of NSCLC cells.
Similar content being viewed by others
Introduction
Lung cancer (LC) is a common malignancy, and non-small cell lung cancer (NSCLC) occupies approximately 85% of LC cases1. The currently available therapies for NSCLC patients mainly include surgical resection, chemotherapy, and radiotherapy2. Radiotherapy is applicable to almost all stages of LC, and cure or alleviate over half of the patients who receive radiotherapy at least once3. Despite the tremendous progress in radiotherapy for NSCLC, the overall cure rate and survival rate of NSCLC are still unfavorable4. Radioresistance is regarded as the major contributor to the high mortality of NSCLC5. Therefore, clarifying the mechanisms behind radioresistance and exploring valid therapeutic targets remain urgent issues to be solved.
Tumorigenesis is correlated with the interference of various critical molecular pathways and biological processes, such as ubiquitination6. Ubiquitination participates in proteasome degradation through the ubiquitin-proteasome system, which is a crucial eukaryotic proteolysis mechanism of proteins implicated in cell cycle, differentiation, and apoptosis7. In the process of ubiquitination, the binding of ubiquitin-protein to the target protein requires the successive action of ubiquitin-activating enzyme (E1), ubiquitin-binding enzyme (E2), and ubiquitin ligase (E3)6. FBXW7 is the main component of Skp1-Cullin1-F-box (SCF)-type ubiquitin ligase8. Importantly, FBXW7 is viewed as a potent tumor suppressor through the degradation of various oncoproteins such as cyclin E, notch, c-Jun, and c-Myc9,10. Targeting FBXW7 may be a promising anti-tumor approach due to the presence of inactivation or deletion of FBXW7 in human malignancies11. FBXW7 can inhibit the epithelial-mesenchymal transition (EMT) and alleviate chemotherapy resistance of NSCLC cells through ubiquitin-dependent degradation12. An increased FBXW7 level is related to the enhanced sensitivity of NSCLC cells to adriamycin13. Higher FBXW7 expression also indicates that patients with advanced esophageal squamous cell carcinoma have good response to chemoradiation therapy14. However, the exact mechanism of FBXW7 in NSCLC radioresistance remains unclear.
SOX9 is the target of E3 ligase FBXW7 for proteasome degradation15. SOX9 is a transcription factor that regulates stem cell proliferation, differentiation, and survival16,17. SOX9 overexpression alone or combination with oncoproteins can initiate the progression of various tumors, including NSCLC18. Upregulated SOX9 expression is related to the histological stage of NSCLC, and patients with higher SOX9 expression exhibit shorter survival19. According to the prediction analysis of the database, Sox9 is associated with a poor prognosis of NSCLC and is highly expressed in NSCLC. Therefore, we chose Sox9 as the research object downstream of FBXW7. Whether E3 ubiquitin ligase FBXW7 can improve the radiosensitivity of NSCLC by regulating SOX9 remains unknown. This study sought to explore the function of FBXW7 in the radiosensitivity of NSCLC, which shall render novel insights into the clinical intervention of NSCLC.
Materials and methods
Construction of radiotherapy resistant cell line
The radioresistant cell model was established using NCI-H1299 cells. When NCI-H1299 cells in the logarithmic phase reached 80–90% confluence, they were irradiated with 6 MV X-ray. The radiation conditions were: 6 MV X-ray generated by a linear accelerator, 10 × 10 cm (radiation field), 200 cGy/min (dose rate), 100 cm (source-skin distance), 200 cGy/min (absorbed dose rate). A sublethal dose was utilized for model establishment. NCI-H1299 cells were subjected to 0, 2, 4, 6, 8, and 10 Gy X-ray radiation. Afterwards, the medium was refreshed and cells were passaged for 5–7 days. The cell growth was observed to determine the sublethal dose. Then, H1299 cells were subjected to sublethal radiation and the surviving cells were cultured to the 1st passage subline cells. Following once sublethal radiation again, the surviving cells were incubated to subline cells of the next passage and received another sublethal irradiation when the cells were in a stable state. After 5 or more regimens of sublethal radiation, NCI-H1299R cells were obtained.
Cell transfection
Parental and resistant NCI-H1299 cells were detached with trypsin and prepared into cell suspensions. The number of cells was calculated and the cell density was adjusted. Next, cells were seeded in the 25 cm2 flask in an incubator at 37 °C with 5% CO2 and transfected until 30–40% confluence.
FBXW7 overexpression plasmid (oe-FBXW7), SOX9 overexpression plasmid (oe-SOX9), CDKN1A shRNA (sh-CDKN1A), and negative controls were supplied by GenePharma (Shanghai, China). Cells transfection was conducted by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h, NCI-H1299 and NCI-H1299R cells were collected for subsequent experiments.
Colony formation assay
Cells were detached with trypsin and then counted. Different numbers of NCI-H1299 and NCI-H1299R cells were seeded into the dish and received different doses of radiation. After cell adherence, they were treated with 0, 2, 4, and 6 Gy X-ray and cultured for 14 days, and then the medium was removed. Afterwards, cells were rinsed twice with PBS, fixed for 15 min in methanol, and stained for 15 min with Giemsa, with the residual dye rinsed with double distilled water. The clones with over 50 cells were regarded as effective cell clones, and the cell survival fraction under different radiations was calculated. Survival fraction = the number of clonal formation in the radiation group of a certain dose/the number of clonal formation in the non-radiation group × 100%.
Cell counting kit-8 (CCK-8) assay
NCI-H1299 and NCI-H1299R cells in the logarithmic phase and transfected cells were detached with trypsin and prepared into cell suspensions. The cells were plated in 96-well plates at 92,000 cells/100 μL, with each well supplemented with 100 μL medium containing serum. Each cell had five duplicated wells. A group of cells with additional culture medium were used as the blank group. After 12 h-incubation, the cells were irradiated with 0, 2, 4, and 6 Gy X-ray, and then the fresh medium was replaced, and cells were routinely cultured. After radiation for 2, 4, 6, and 8 days, 10 μL CCK-8 was supplemented for 0.5 h-incubation. The OD at 450 nm was measured by the microplate reader. With the OD value representing the number of cells, the survival curves of NCI-H1299 and NCI-H1299R cells were drawn.
Flow cytometry
Cells were irradiated at a dose of 0, 2, 4, and 6 Gy, respectively. The medium was refreshed after radiation. After 24 h of conventional culture, the cell medium was transferred into a 15 mL centrifuge tube. Cells were rinsed with PBS and detached with an appropriate amount of trypsin. Trypsin reaction was terminated by adding the medium containing serum. The collected medium was centrifuged for 5 min at 1000 r/min, rinsed with 1 mL PBS, and counted (no less than 1 × 105). After centrifugation at 1000 rpm for 5 min, the cells were suspended in 1 × binding buffer (400 μL) and mixed with Annexin V-FITC (5 μL), followed by incubation under dark conditions for 15 min. Subsequently, cells were added with 10 μL PI, bathed on ice, and measured on a flow cytometer.
Immunofluorescence
The coverslips placed in 6-well plates were coated with polyethyleneimine (0.04% v/v, Sigma-Aldrich, St. Louis, MO, USA). The cells in each group were irradiated at a dose of 6 Gy and seeded onto the coverslips containing medium for routine culture. When the cells reached 80–90% confluence, they were fixed with 4% paraformaldehyde. Next, the coverslips were incubated with rabbit monoclonal antibody γ-H2AX (1:250, ab81299, Abcam, Cambridge, UK) in a damp dark room at 4 °C for 1 h, rinsed with 0.1% Triton-X and 1 × PBS (PBST), and treated with donkey anti-rabbit IgG (Alexa Fluor® 647) (ab150075, 1:200, Abcam) for 1 h, following washing with PBST three times (5 min/time). Afterwards, the coverslips were incubated with 4', 6-diamino-2-phenylindole for 10 min to counterstain the nuclei and sealed on the slides with Vectashield fluorescent mounting medium (H-1000, Sangerbio, Shanghai, China). The images were obtained by Olympus Life Sciences (Japan). The γ-H2AX foci were counted through Image J software (National Institutes of Health, Bethesda, Maryland, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted by RNeasy Mini kits (Qiagen, Valencia, CA, USA) and reverse transcribed into cDNA using reverse transcription kits (RR047A, Takara, Toyko, Japan). qRT-PCR was performed using SYBR® Premix Ex TaqTM II kit (DRR081, Takara) on a real-time qPCR instrument (7500, ABI, Foster City, CA, USA). The primers (Sangon Biotech) are exhibited in Table 1. Ct value was recorded and gene expression was calculated by the 2−ΔΔCt method, with GAPDH as an internal reference.
Western blot
The cells or tissues were lysed in the enhanced RIPA lysate containing protease inhibitor (Boster, Wuhan, China), and the protein concentration was determined by the bicinchoninic acid (BCA) kit (Boster). Subsequently, the protein was isolated by 10% SDS-PAGE, transferred onto PVDF membranes, blocked for 2 h with 5% BSA, and incubated with diluted primary antibodies rabbit anti-FBXW7 (1:1000, ab192328, Abcam), SOX9 (ab185966, 1:1000, Abcam) and GAPDH (ab9485, 1:2500, Abcam) at 4 °C overnight. Then the membranes were probed for 1 h with HRP-labeled goat anti-rabbit IgG (ab205718, 1:2000, Abcam). The bands were developed with enhanced chemiluminescence (Millipore, Billerica, MA, USA). The gray value was calculated using Image Pro Plus 6.0 (Media Cybernetics, San Diego, CA, USA).
Immunoprecipitation
Cells were immersed in lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 10% glycerol, 1 mM EDTA, 0.5% NP-40, and protease inhibitor]. The cell debris was discarded by centrifugation. The lysate concentration was examined by BCA kits, and then 1 μg FBXW7 antibody (ab109617, Abcam) was supplemented with 500 μg protein and rotated gently at 4 °C overnight. Afterwards, 40 μL protein A + G agarose beads (Beyotime, Shanghai, China) were added to capture the immune complex, and then the mixture was gently rotated at 4 °C for 4 h and centrifuged for 5 min at 1000 × g and 4 °C to discard the supernatant. The precipitate was then washed five times with cold PBS and boiled in SDS sample buffer for 5 min to dissociate the immune complex from the magnetic beads. Finally, the supernatant was collected by centrifugation for western blot analysis.
Ubiquitin pull-down
NCI-H1299 cells were transfected with oe-NC or oe-FBXW7, and total cell lysates were collected by the above method. An equal amount of SOX9 antibody (ab185230, 1:60, Abcam) was added to 500 g protein, and the mixture was gently rotated overnight at 4 °C. Then, 40 μL protein A + G agarose beads (Beyotime) were added and incubated at 4 °C for 4 h to capture the immune complex. After centrifugation at 4 °C for 5 min, the supernatant was discarded. The precipitate was washed five times with cold PBS and boiled in SDS sample buffer for 5 min to dissociate the immune complex from the magnetic beads. Immunoprecipitation was analyzed by western blotting with an anti-ubiquitin antibody (ab134953, 1:2000, Abcam).
Determination of SOX9 protein stability of cycloheximide (CHX) and MG132
NCI-H1299 cells transfected with oe-NC or oe-FBXW7 were treated with 10 μM CHX at the designated time (0, 8, 16, and 24 h) to block protein synthesis. Besides, NCI-H1299 cells transfected with oe-NC or oe-FBXW7 were treated with 20 μM MG132 to inhibit proteasome before collection. The protein was extracted and the SOX9 level was examined by western blot.
Dual-luciferase reporter gene assay
The binding site of SOX9 protein in the CDKN1A promoter was analyzed by UCSC (http://genome.ucsc.edu/) and JASPAR websites (http://jaspar.genereg.net/). The recombinant luciferase reporter gene vectors of wild-type (CDKN1A-WT) and mutant-type (CDKN1A-MUT) sites were constructed and co-transfected with SOX9 expression vector into HEK-293T cells (Cell Resource Center, Shanghai Institutes for Biological Sciences, the Chinese Academy of Sciences). oe-NC and oe-SOX9 were co-transfected with CDKN1A-WT and CDKN1A-MUT plasmids respectively to examine whether SOX9 was bound to the CDKN1A promoter. After 48 h, the luciferase activity was determined by luciferase assay kits (K801-200, Biovision, San Francisco, USA) and Glomax20/20 luminometer (Promega, Madison, WI, USA).
Chromatin immunoprecipitation (ChIP)
Cells were fixed with formaldehyde for 10 min to produce DNA-protein crosslinking. The ultrasonic crusher was set as 10 s/time, with an interval of 10 s and 15 cycles to break the cells and the chromatin into fragments. Then cells were treated with antibodies against IgG (ab172730, 1:1000, Abcam) and SOX9 (ab185230, 1:60, Abcam) at 4 °C overnight. DNA-protein complexes were precipitated by Protein Agarose/Sepharose. After centrifugation at 12,000 × g for 5 min, the supernatant was discarded. The nonspecific complexes were washed and decrosslinked at 65 °C overnight. DNA fragment was extracted and purified by phenol/chloroform extraction, and the binding of SOX9 was detected by qRT-PCR with CDKN1A specific primer.
Xenograft tumor in nude mice
BALB/c nude mice (4-6 weeks old) were purchased from Beijing Vital River Laboratory. Lentivirus LV-oe-NC and LV-oe-FBXW7 were purchased from GenePharma. NCI-H1299 cells and NCI-H1299R cells were infected with LV-oe-NC and LV-oe-FBXW7, respectively. Then, 2 × 106 cells were suspended in 20 μL PBS and subcutaneously injected into the hind legs of nude mice (N = 12). The diameter of the tumor was observed and measured. When the tumor diameter reached 0.5–1.0 cm, the mouse received 6 Gy X-ray radiation every 3 days, totally 3 times20. After 3 times of radiation, the mice were euthanized by injecting excessive pentobarbital sodium (i.p, 100 mg/kg). The subcutaneous tumor was removed and tumor volume and weight were measured. Tumor volume (V) = 0.5 × L × W2, where L and W were defined as tumor length and width. Then 6 rats in each group were used for RT-qPCR and western blot experiments, and the other 6 tumors were sliced after embedding and stained with TUNEL.
TUNEL staining
According to the instructions of TUNEL detection kits (No. 11684817910; Roche Diagnostics GmbH), paraffined sections were dewaxed, hydrated, and incubated with protease K working solution at room temperature. After washing with PBS, the sections were placed in TUNEL reaction mixture and incubated under dark conditions for 1 h. All sections were analyzed by Image J software under the microscope, the visual field was randomly selected, and the number of positive cells was counted.
Statistical analysis
Data were processed using SPSS 21.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 8.0 (GraphPad Inc., San Diego, CA, USA). Data are described as mean ± standard deviation. The t test was utilized for two-group comparisons. One-way or two-way analysis of variance (ANOVA) was adopted for multiple group comparisons, followed by Tukey’s multiple comparison test. p < 0.05 meant significant difference.
Results
Resistant cells were more resistant to radiation than parental cells
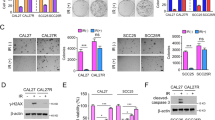
Parental NCI-H1299 cells were used to construct resistant NCI-H1299R cells. NCI-H1299 and NCI-H1299R cells were irradiated with 0, 2, 4, and 6 Gy X-ray for 14 days. Then the cell survival was detected using colony formation assay. The results demonstrated that compared with that of NCI-H1299R cells, the number of cell clones of NCI-H1299 cells was decreased after exposure to gradient doses of radiation (2, 4, and 6 Gy), and the ability to inhibit colony formation was more effective at 6 Gy X-ray (Fig. 1A). The proliferation curve and apoptosis of NCI-H1299 and NCI-H1299R cells were detected using CCK-8 assay and flow cytometry, respectively. Compared with NCI-H1299R cells, NCI-H1299 cells exhibited reduced proliferation under radiation (Fig. 1B, p < 0.05) and notably increased apoptosis rates (Fig. 1C, p < 0.05). Moreover, after 6 Gy X-ray radiation, immunofluorescence detection of γ-H2AX (a kind of DNA double-strand damage marker; When DNA is damaged by radiation, the double strand breaks and forms γ-H2AX, so we can intuitively observe the degree of DNA damage of cells by detectingγ-H2AX) showed that the number of γ-H2AX-positive foci in NCI-H1299 cells was notably increased (Fig. 1D, p < 0.05), suggesting that DNA damage was aggravated. Taken together, NCI-H1299R was more resistant to radiation than NCI-H1299 cells.
A NCI-H1299 and NCI-H1299R cells were irradiated with 0, 2, 4, and 6 Gy dose of X-ray for 14 days, respectively, and then the cell survival was detected using colony formation assay. B, C NCI-H1299 and NCI-H1299R cells were further irradiated with 6 Gy dose of X-ray, and the proliferation and apoptosis were detected using CCK-8 assay and flow cytometry. D γ-H2AX was detected using immunofluorescence, scale bars: 10 μm. The cell experiment was conducted 3 times independently. Data were presented as mean ± standard deviation. Data in A/B were analyzed using two-way ANOVA, and data in C/D were analyzed using t test, followed by Tukey’s multiple comparison test, *p < 0.05.
FBXW7 was depressed in radioresistant cells, and overexpression of FBXW7 enhanced radiosensitivity
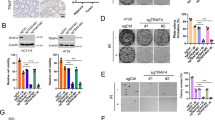
FBXW7 is implicated in cisplatin resistance in NSCLC21, but the role of FBXW7 in radioresistance in NSCLC remains unknown. GEPIA (http://gepia.cancer-pku.cn/detail.php?gene=FBXW7) and Kaplan-Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=lung) website exhibited that FBXW7 was under-expressed in LC (Fig. 2A) and concerned with unfavorable outcomes of squamous cell carcinoma and adenocarcinoma (Fig. 2B), implying that FBXW7 may modulate the initiation and progression of NSCLC. Additionally, FBXW7 was notably lowered in NCI-H1299R cells relative to that in NCI-H1299 cells (Fig. 2C, D, p < 0.05).
A FBXW7 was poorly expressed in LC (including squamous cell carcinoma and adenocarcinoma) through GEPIA website (http://gepia.cancer-pku.cn/detail.php?gene=FBXW7). In the figure, red is the tumor group and gray is the normal group. B FBXW7 low expression was associated with the poor prognosis of LC (including squamous cell carcinoma and adenocarcinoma) through Kaplan-Meier Plotter website (http://kmplot.com/analysis/index.php?p=service&cancer=lung). C, D FBXW7 expression in NCI-H1299 and NCI-H1299R cells was detected using qRT-PCR and western blot. E, F NCI-H1299 and NCI-H1299R cells were transfected with oe-NC and oe-FBXW7, and then FBXW7 expression was detected using qRT-PCR and western blot. G, H NCI-H1299 and NCI-H1299R cells transfected with oe-NC and oe-FBXW7 were irradiated with 6 Gy dose of X-ray, and then cell proliferation was measured using CCK-8 and colony formation assay. I γ-H2AX was detected using immunofluorescence, scale bars: 10 μm. The cell experiment was conducted 3 times independently. Data were presented as mean ± standard deviation. Data in E–I were analyzed using two-way ANOVA, and data in C/D were analyzed using t test, followed by Tukey’s multiple comparison test, *p < 0.05.
Next, NCI-H1299 cells and NCI-H1299R cells were transfected with oe-FBXW7 (Fig. 2E, F, p < 0.05) to study the effect of FBXW7 on the radiosensitivity of cells. The results exhibited that overexpression of FBXW7 significantly repressed the proliferation and colony formation of NCI-H1299 and NCI-H1299R cells after 6 Gy X-ray radiation (Fig. 2G, H, p < 0.05) and increased the number of γ-H2AX-positive foci (Fig. 2I, p < 0.05). Briefly, FBXW7 augmented the radiosensitivity of NSCLC cells.
SOX9 could be ubiquitinated by FBXW7 and degraded by proteasome
SOX9 gene encodes transcription factors that control cell fate throughout tissue homeostasis. SOX9 may lead to unfavorable prognoses by increasing therapy resistance in tumors19. GEPIA website (http://gepia.cancer-pku.cn/detail.php?gene=SOX9) exhibited that SOX9 was elevated in LC (including squamous cell carcinoma and adenocarcinoma) (Fig. 3A) and correlated with unfavorable outcomes (Fig. 3B). In NCI-H1299 and NCI-H1299R cells transfected with oe-FBXW7, overexpression of FBXW7 did not affect the mRNA expression of SOX9, but significantly reduced the protein level of SOX9 (Fig. 3C, D, p < 0.05), suggesting that FBXW7 might regulate SOX9 expression at the protein level.
A, B GEPIA website (http://gepia.cancer-pku.cn/detail.php?gene=SOX9) showed that SOX9 was highly expressed in LC (including squamous cell carcinoma and adenocarcinoma) and associated with poor prognosis. In the figure, red is the tumor group and gray is the normal group. C, D NCI-H1299 and NCI-H1299R cells were transfected with oe-NC and oe-FBXW7, and then SOX9 expression was detected using qRT-PCR and western blot. E The interaction between FBXW7 and SOX9 was detected by immunoprecipitation assay in NCI-H1299 cells. F Ubiquitin pull-down assay was used to detect the ubiquitin level of SOX9 after transfection of oe-NC and oe-FBXW7 into NCI-H1299 cells. G The transfected NCI-H1299 cells were treated with CHX (10 μM) for 0, 8, 16, and 24 h to inhibit protein synthesis, and then SOX9 expression was detected using western blot. H The transfected NCI-H1299 cells were treated with MG132 (20 μM) for 8 h, and then SOX9 expression was detected using western blot. The cell experiment was conducted 3 times independently. Data were presented as mean ± standard deviation and analyzed using two-way ANOVA, followed by Tukey’s multiple comparison test, *p < 0.05.
SOX9 can be ubiquitinated and degraded by proteasome and is associated with DNA damage15. Immunoprecipitation assay was performed in NCI-H1299 cells, and the results revealed that FBXW7 could interact with SOX9 (Fig. 3E). Ubiquitin pull-down assay demonstrated that overexpression of FBXW7 in NCI-H1299 cells increased the ubiquitination level of SOX9 (Fig. 3F). CHX test showed that the degradation rate of SOX9 protein was significantly accelerated after overexpression of FBXW7 (Fig. 3G, p < 0.05), while the protein level of SOX9 was notably increased after the addition of proteasome inhibitor MG132 (Fig. 3H, p < 0.05). Taken together, FBXW7 interacted with SOX9 and reduced the stability of SOX9 protein by mediating the ubiquitination degradation of SOX9, thus suppressing the protein level of SOX9.
FBXW7 enhanced the radiosensitivity of NSCLC cells by inhibiting SOX9 expression
Based on the above results, we speculated that the inhibitory effect of FBXW7 on radioresistance of NSCLC was realized by regulating SOX9. We overexpressed FBXW7 and SOX9 in NCI-H1299R cells. Western blot demonstrated that compared with the oe-FBXW7 + oe-NC group, the oe-FBXW7 + oe-SOX9 group showed notably increased SOX9 expression (Fig. 4A, p < 0.05). CCK-8 assay showed that overexpression of SOX9 promoted the proliferation of NCI-H1299R cells after 6 Gy X-ray radiation (Fig. 4B, p < 0.05) and reduced the number of γ-H2AX-positive foci (Fig. 4C, p < 0.05). In brief, FBXW7 improved the radiosensitivity by inhibiting SOX9 expression.
FBXW7 and SOX9 were overexpressed in NCI-H1299R cells. A SOX9 expression was detected using western blot. B CCK-8 assay was used to detect the cell proliferation curve after 6 Gy dose of X-ray radiation. C γ-H2AX was detected using immunofluorescence, scale bars: 10 μm. The cell experiment was conducted 3 times independently. Data were presented as mean ± standard deviation. Data in A/C were analyzed using one-way ANOVA, and data in B were analyzed using two-way ANOVA, followed by Tukey’s multiple comparison test, *p < 0.05.
FBXW7 promoted CDKN1A expression via SOX9
As a transcription factor, SOX9 can repress the promoter activity of target genes22. To identify the downstream mechanism of SOX9, we consulted the literature and noticed that SOX9 was negatively correlated with CDKN1A in adenocarcinoma, but the regulatory mechanism between them has not been elucidated in depth23. GEPIA website (http://gepia.cancer-pku.cn/detail.php?gene=CDKN1A) demonstrated that CDKN1A was downregulated in LC (including squamous cell carcinoma and adenocarcinoma) (Fig. 5A) and correlated with the prognosis of LC (Fig. 5B), suggesting the implication of CDKN1A in the initiation and progression of NSCLC. Subsequent qRT-PCR showed that overexpression of FBXW7 significantly enhanced CDKN1A gene expression, while oe-SOX9 reversed the effect of oe-FBXW7 on CDKN1A expression (Fig. 5C, p < 0.05), suggesting that FBXW7 promoted CDKN1A expression by inhibiting SOX9.
A, B GEPIA website (http://gepia.cancer-pku.cn/detail.php?gene=SOX9) showed that CDKN1A was highly expressed in LC (including squamous cell carcinoma and adenocarcinoma) and associated with poor prognosis. In the figure, red is the tumor group and gray is the normal group. C CDKN1A expression in NCI-H1299R cells was detected using qRT-PCR. D JASPAR website (http://jaspar.genereg.net/analysis) predicted that SOX9 had a binding site in the promoter of CDKN1A. E, F Dual-luciferase reporter gene assay and ChIP assay verified that SOX9 could bind to the promoter of CDKN1A. The cell experiment was conducted 3 times independently. Data were presented as mean ± standard deviation. Data in C were analyzed using one-way ANOVA, data in E were analyzed using two-way ANOVA, and data in F were analyzed using t test, followed by Tukey’s multiple comparison test, *p < 0.05.
JASPAR website (http://jaspar.genereg.net/analysis) predicted that SOX9 had a binding site in the CDKN1A promoter (Fig. 5D). Dual-luciferase assay verified that overexpression of SOX9 significantly reduced the luciferase activity of CDKN1A-WT (Fig. 5E, p < 0.05). ChIP assay in NCI-H1299 cells also revealed that SOX9 could bind to the promoter region of CDKN1A (Fig. 5F, p < 0.05). Overall, SOX9 inhibited by FBXW7 could bind to the CDKN1A promoter as a transcription factor, thus inhibiting CDKN1A expression.
FBXW7 enhanced the radiosensitivity of NSCLC cells by increasing CDKN1A expression
Next, we conducted functional rescue experiments to verify the above regulatory mechanisms. NCI-H1299R cells were transfected with oe-FBXW7 and sh-CDKN1A. CDKN1A expression in the oe-FBXW7 + sh-CDKN1A group was notably lower than that the oe-FBXW7 + sh-NC group (Fig. 6A, p < 0.05). After 6 Gy X-ray radiation, silencing CDKN1A promoted the proliferation of NCI-H1299R cells (Fig. 6B, p < 0.05) and reduced the number of γ-H2AX-positive foci (Fig. 6C, p < 0.05). Altogether, FBXW7 augmented radiosensitivity of NSCLC by elevating CDKN1A expression.
NCI-H1299R cells were transfected with oe-FBXW7 and sh-CDKN1A. A CDKN1A expression was detected using qRT-PCR. B CCK-8 assay was used to detect the cell proliferation curve after 6 Gy dose of X-ray radiation. C γ-H2AX was detected using immunofluorescence, scale bars: 10 μm. The cell experiment was conducted 3 times independently. Data were presented as mean ± standard deviation. Data in A/C were analyzed using one-way ANOVA, and data in B were analyzed using two-way ANOVA, followed by Tukey’s multiple comparison test, *p < 0.05.
FBXW7 augmented radiosensitivity of NSCLC cells in vivo via SOX9/CDKN1A
To verify the effect of FBXW7 on the radiosensitivity of cells in vivo, we infected NCI-H1299 cells and NCI-H1299R cells with LV-oe-NC and LV-oe-FBXW7 respectively and established xenograft tumor models. When the tumor diameter reached 0.5–1.0 cm, the mice received 6 Gy X-ray. Compared with the LV-oe-NC group, the LV-oe-FBXW7 group showed reduced tumor volume and weight (Fig. 7A–C, p < 0.05), increased FBXW7 and CDKN1A levels (Fig. 7D, E, p < 0.05), decreased SOX9 protein level (Fig. 7F, p < 0.05), and increased apoptosis (Fig. 7G, p < 0.05). Briefly, FBXW7 inhibited tumorigenesis in vivo and improved radiosensitivity by inhibiting SOX9 and promoting CDKN1A expression.
NCI-H1299 cells and NCI-H1299R cells were infected with LV-oe-NC and LV-oe-FBXW7 respectively to establish the subcutaneous xenograft tumor model. When the tumor diameter reached 0.5–1.0 cm in nude mice, the mice were irradiated with 6 Gy X-ray. A Images of tumors. B Tumor volume. C Tumor weight. D, E FBXW7 and CDKN1A expression was detected using qRT-PCR. F SOX9 expression was detected using western blot. G Apoptosis in the tumor was detected by TUNEL staining. N = 6. Data were presented as mean ± standard deviation and analyzed using two-way ANOVA, followed by Tukey’s multiple comparison test, *p < 0.05.
Discussion
Radiotherapy combined with chemotherapy and/or surgery can exhibit synergistic effects on NSCLC treatment5. Unfortunately, the efficacy of radiotherapy is largely limited by acquired resistance, eventually leading to tumor recurrence and metastasis24. E3 ubiquitin ligase plays a vital role in tumor initiation and progression by facilitating protein ubiquitination and degradation, thus offering a potential therapeutic option25. FBXW7 is a member of the E3 ubiquitin ligases and targets the network of oncoproteins for degradation7. This study demonstrated the underlying mechanism of FBXW7 in the radiosensitivity of NSCLC cells.
FBXW7 modulates the network of critical oncoproteins26. FBXW7 knockdown has been demonstrated to facilitate EMT of NSCLC cells and confer chemotherapy resistance27. Nevertheless, the effect of FBXW7 on the radioresistance of NSCLC remains unknown. In this study, the parental NCI-H1299 cells were used to construct radioresistant NCI-H1299R cells, and the results revealed that NCI-H1299R cells were more resistant to radiation than NCI-H1299 cells. GEPIA website showed that FBXW7 was under-expressed in LC and associated with unfavorable outcomes, implying the involvement of FBXW7 in NSCLC. We demonstrated that FBXW7 expression was notably reduced in NCI-H1299R cells. Then NCI-H1299 cells and NCI-H1299R cells were transfected with oe-FBXW7 to verify the effect of FBXW7 on the radiosensitivity of cells. Overexpression of FBXW7 significantly repressed the cell proliferation and increased the γ-H2AX-positive foci with irradiation. To our knowledge, we were the first to reveal that FBXW7 improved the radiosensitivity of NSCLC cells. SOX9 is a substrate of FBW7, and FBW7 depletion results in SOX9 stabilization15. Upregulation of SOX9 can contribute to tumor progression and therapy resistance, thus leading to poor survival18. Elevated SOX9 expression induces EMT and facilitates distant metastasis in NSCLC via the Wnt/β-catenin pathway28. The FBW7-SOX9 axis is implicated in therapy resistance in cancer29. GEPIA website exhibited that SOX9 was elevated in LC and concerned with unfavorable prognoses. FBXW7 overexpression did not affect the mRNA expression of SOX9, but significantly reduced the protein level of SOX9, suggesting that FBXW7 modulated SOX9 expression at the protein level. This study further revealed that FBXW7 interacted with SOX9 and reduced the stability of SOX9 protein by mediating the ubiquitination degradation of SOX9, thus suppressing the protein level of SOX9. Next, we overexpressed FBXW7 and SOX9 expression in NCI-H1299R cells and confirmed that FBXW7 improved the radiosensitivity by inhibiting SOX9 expression.
Subsequently, we focused on the downstream mechanism of SOX9. SOX9 serves as a hallmark of lung adenocarcinoma and contributes to tumor growth by negatively affecting the cell cycle regulator CDKN1A23. CDKN1A is initially identified as an inhibitor of cell proliferation and DNA replication, which has become a target of anti-tumor therapy30. Emerging evidence has unveiled that CDKN1A knockdown may contribute to chemotherapy resistance of NSCLC31,32,33. Whether CDKN1A is involved in the radiosensitivity of NSCLC needs further exploration. GEPIA website demonstrated that CDKN1A was downregulated in LC. Overexpression of FBXW7 significantly enhanced CDKN1A expression, while overexpression of SOX9 reversed the effect of FBXW7 overexpression on CDKN1A expression, indicating that FBXW7 promoted CDKN1A expression by inhibiting SOX9. Dual-luciferase reporter gene assay and ChIP assay verified that SOX9 could bind to the CDKN1A promoter. In brief, SOX9 inhibited by ubiquitination of FBXW7 could bind to the CDKN1A promoter as a transcription factor, thus inhibiting CDKN1A expression. Functional rescue experiments were conducted to verify the above regulatory mechanisms. NCI-H1299R cells were transfected with oe-FBXW7 and sh-CDKN1A. The results confirmed that FBXW7 enhanced the radiosensitivity of NSCLC cells by promoting CDKN1A expression. Moreover, we infected NCI-H1299 cells and NCI-H1299R cells with LV-oe-NC and LV-oe-FBXW7 respectively to establish xenograft tumor models. In vivo experiments also validated that FBXW7 promoted CDKN1A expression by inhibiting SOX9, thereby suppressing tumorigenesis and improving radiosensitivity.
To sum up, E3 ubiquitin ligase FBXW7 enhances the radiosensitivity of NSCLC by suppressing the binding of SOX9 to CDKN1A and upregulating CDKN1A expression (Fig. 8). This study may hint at a promising target for improving the radiosensitivity of NSCLC. However, this study failed to explore the deeper regulatory mechanism of abnormal expression of FBXW7 in radioresistant cells and other ubiquitinated proteins downstream of FBXW7. In the future, we shall further elucidate the mechanism involved in the abnormal expression of FBXW7 in radioresistant cells and conduct more in-depth research on other ubiquitinated proteins downstream of FBXW7 to explore the regulatory mechanism of FBXW7 in the radiosensitivity of NSCLC.
Data availability
The data that support this study are available from the corresponding author upon reasonable request.
References
Burdett S, Rydzewska L, Tierney J, Fisher D, Parmar MK, Arriagada R et al. Postoperative radiotherapy for non-small cell lung cancer. Cochrane Database Syst Rev 10, CD002142 (2016)
Osarogiagbon RU, Lin CC, Smeltzer MP, Jemal A. Prevalence, Prognostic Implications, and Survival Modulators of Incompletely Resected Non-Small Cell Lung Cancer in the U.S. National Cancer Data Base. J Thorac Oncol 11, e5-16 (2016)
Brown S, Banfill K, Aznar MC, Whitehurst P, Faivre Finn C. The evolving role of radiotherapy in non-small cell lung cancer. Br J Radiol 92, 20190524 (2019)
Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 553, 446-454 (2018)
Yun HS, Baek JH, Yim JH, Um HD, Park JK, Song JY et al. Radiotherapy diagnostic biomarkers in radioresistant human H460 lung cancer stem-like cells. Cancer Biol Ther 17, 208-218 (2016)
Sailo BL, Banik K, Girisa S, Bordoloi D, Fan L, Halim CE et al. FBXW7 in Cancer: What Has Been Unraveled Thus Far? Cancers (Basel) 11 (2019)
Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 26, 455-464 (2014)
Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer 17, 115 (2018)
Jardim DL, Wheler JJ, Hess K, Tsimberidou AM, Zinner R, Janku F et al. FBXW7 mutations in patients with advanced cancers: clinical and molecular characteristics and outcomes with mTOR inhibitors. PLoS One 9, e89388 (2014)
Zhou Z, He C, Wang J. Regulation mechanism of Fbxw7-related signaling pathways (Review). Oncol Rep 34, 2215-2224 (2015)
Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget 5, 2000-2015 (2014)
Xiao G, Li Y, Wang M, Li X, Qin S, Sun X et al. FBXW7 suppresses epithelial-mesenchymal transition and chemo-resistance of non-small-cell lung cancer cells by targeting snai1 for ubiquitin-dependent degradation. Cell Prolif 51, e12473 (2018)
Li R, Wu S, Chen X, Xu H, Teng P, Li W. miR-223/FBW7 axis regulates doxorubicin sensitivity through epithelial mesenchymal transition in non-small cell lung cancer. Am J Transl Res 8, 2512-2524 (2016)
Gombodorj N, Yokobori T, Tanaka N, Suzuki S, Kuriyama K, Kumakura Y et al. Correlation between high FBXW7 expression in pretreatment biopsy specimens and good response to chemoradiation therapy in patients with locally advanced esophageal cancer: A retrospective study. J Surg Oncol 118, 101-108 (2018)
Hong X, Liu W, Song R, Shah JJ, Feng X, Tsang CK et al. SOX9 is targeted for proteasomal degradation by the E3 ligase FBW7 in response to DNA damage. Nucleic Acids Res 44, 8855-8869 (2016)
Larsimont JC, Youssef KK, Sanchez-Danes A, Sukumaran V, Defrance M, Delatte B et al. Sox9 Controls Self-Renewal of Oncogene Targeted Cells and Links Tumor Initiation and Invasion. Cell Stem Cell 17, 60-73 (2015)
Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366-370 (2015)
Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res 72, 1301-1315 (2012)
Zhou CH, Ye LP, Ye SX, Li Y, Zhang XY, Xu XY et al. Clinical significance of SOX9 in human non-small cell lung cancer progression and overall patient survival. J Exp Clin Cancer Res 31, 18 (2012)
Yu Z, Wang G, Zhang C, Liu Y, Chen W, Wang H et al. LncRNA SBF2-AS1 affects the radiosensitivity of non-small cell lung cancer via modulating microRNA-302a/MBNL3 axis. Cell Cycle 19, 300-316 (2020)
Song Y, Liu Y, Pan S, Xie S, Wang ZW, Zhu X. Role of the COP1 protein in cancer development and therapy. Semin Cancer Biol 67, 43-52 (2020)
Qi J, Yang Y, Hao P, Xu J. Transcription Factor SOX9 Promotes Osteosarcoma Cell Growth by Repressing Claudin-8 Expression. Tohoku J Exp Med 241, 55-63 (2017)
Jiang SS, Fang WT, Hou YH, Huang SF, Yen BL, Chang JL et al. Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clin Cancer Res 16, 4363-4373 (2010)
Ozpiskin OM, Zhang L, Li JJ. Immune targets in the tumor microenvironment treated by radiotherapy. Theranostics 9, 1215-1231 (2019)
Li X, Elmira E, Rohondia S, Wang J, Liu J, Dou QP. A patent review of the ubiquitin ligase system: 2015-2018. Expert Opin Ther Pat 28, 919-937 (2018)
Cao J, Ge MH, Ling ZQ. Fbxw7 Tumor Suppressor: A Vital Regulator Contributes to Human Tumorigenesis. Medicine (Baltimore) 95, e2496 (2016)
Xiao Y, Yin C, Wang Y, Lv H, Wang W, Huang Y et al. FBXW7 deletion contributes to lung tumor development and confers resistance to gefitinib therapy. Mol Oncol 12, 883-895 (2018)
Huang JQ, Wei FK, Xu XL, Ye SX, Song JW, Ding PK et al. SOX9 drives the epithelial-mesenchymal transition in non-small-cell lung cancer through the Wnt/beta-catenin pathway. J Transl Med 17, 143 (2019)
Suryo Rahmanto A, Savov V, Brunner A, Bolin S, Weishaupt H, Malyukova A et al. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J 35, 2192-2212 (2016)
Stivala LA, Cazzalini O, Prosperi E. The cyclin-dependent kinase inhibitor p21CDKN1A as a target of anti-cancer drugs. Curr Cancer Drug Targets 12, 85-96 (2012)
Zamagni A, Pasini A, Pirini F, Ravaioli S, Giordano E, Tesei A et al. CDKN1A upregulation and cisplatinpemetrexed resistance in nonsmall cell lung cancer cells. Int J Oncol 56, 1574-1584 (2020)
Xu T, Yan S, Wang M, Jiang L, Ma P, Lu B et al. LncRNA UCA1 Induces Acquired Resistance to Gefitinib by Epigenetically Silencing CDKN1A Expression in Non-small-Cell Lung Cancer. Front Oncol 10, 656 (2020)
Xu S, Huang H, Chen YN, Deng YT, Zhang B, Xiong XD et al. DNA damage responsive miR-33b-3p promoted lung cancer cells survival and cisplatin resistance by targeting p21(WAF1/CIP1). Cell Cycle 15, 2920-2930 (2016)
Funding
The study was sponsored by Urumqi Science and Technology Bureau for the research on Changes of plasma EGFR gene mutation after TKI in advanced non-small cell lung cancer [Y161310003]. The funding organisations had no role in the concept, design, analysis or writing of the manuscript.
Author information
Authors and Affiliations
Contributions
HZ and XW: conceived and designed the experiments, and participated in design and coordination, and helped to draft the manuscript and revise the manuscript. XZ and SL: performed investigation, statistical analysis, and drafted the manuscript. GG and CL: contributed to the acquisition and analysis of data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the Ethical Committee of The Affiliated Tumor Hospital of Xinjiang Medical University (20200288). Animal experiments were conducted under the guidelines of the National Institutes of Health.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhu, H., Wang, X., Zhou, X. et al. E3 ubiquitin ligase FBXW7 enhances radiosensitivity of non-small cell lung cancer cells by inhibiting SOX9 regulation of CDKN1A through ubiquitination. Lab Invest 102, 1203–1213 (2022). https://doi.org/10.1038/s41374-022-00812-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-022-00812-9