Abstract
Self-propelled biomolecular motor systems, such as microtubule-kinesin or microtubule-dynein, are intricate natural machines with the ability to convert chemical energy into mechanical work with high efficiency. In recent years, the biomolecular motor systems have emerged as promising candidates for studying active self-assembly based on the in vitro gliding assay of the motor systems. Several strategies have been developed to demonstrate the active self-assembly of biomolecular motors, which provided a wealth of organized and complex structures. Here we review the latest progress in the active self-assembly of microtubule-kinesin and microtubule-dynein with an emphasis on the emergence of various structures and necessary design parameters for controlling their polymorphism.
Similar content being viewed by others
Introduction
Self-assembly is a highly prevalent phenomenon in nature in which organized and functional structures are spontaneously formed from disordered discrete components [1, 2]. Nature vastly utilizes self-assembly to design various architectures that has significant roles in maintaining necessary functions in living organisms [1]. From a broader perspective, self-assembly can be categorized into two groups: passive and active self-assembly [3, 4]. In passive self-assembly, thermal fluctuations serve as the driving force to move the isolated building blocks through space to form organized structures. Self-assembly in supramolecular systems or conventional fabrication techniques belongs to the category of passive self-assembly. In contrast, active self-assembly is driven by external sources of energy and has an advantage over passive self-assembly in that it is able to surmount the limitation of assembly speed encountered by passive self-assembly and can form complex non-equilibrium structures. It is noteworthy that active self-assembly differs from active self-organization in that self-organization is a process in which the formation of patterns or structures is maintained in a non-equilibrium dynamic state by the continuous flow of energy, whereas active self-assembly uses energy flow in the formation but not in the maintenance of the structures, as the structures are thermodynamically stable. Living organisms are by far the most prominent examples of active self-assembly systems in which hierarchical macroscale structures are formed from molecular components through energy-driven assembly processes [5]. Some of the fascinating examples are cilia and mitotic spindles in living organisms, in which microtubules (MTs), a self-assembled structure of globular protein tubulins, play versatile roles in their organization and functions [6, 7]. By virtue of recent advances in science and technology, it is now possible to reconstruct those cellular components in cell-free conditions in their full functional and structural form. This ability has opened a door to employing biological units in exploring active self-assembly processes in the synthetic world by utilizing their functional advantages over artificial units [8, 9]. A great deal of effort has been dedicated in recent years to studying the self-assembly of biomacromolecules, e.g., MTs and actin in vitro [10,11,12]. A large part of those efforts explored the self-assembly of microtubules and actin in static conditions, i.e., through passive self-assembly [13]. The greatest hurdle in the self-assembly of MTs or actins was controlling the interactions among the building blocks in a user-defined route. The application of dynamic conditions has started to evolve in recent years to overcome this limitation. As a consequence, several methods have been developed that were successful in regulating the interactions among the building blocks and allowed the formation of stable, large-scale self-assembled structures with a much faster rate than that in the passive self-assembly [10]. In the study of the active self-assembly of MTs, a model experimental setup, named the in vitro gliding assay, has been extensively utilized. In this experimental design, biomolecular motors such as kinesin or dynein are adsorbed to the surface of a substrate, which then propel MTs by utilizing the energy obtained from the hydrolysis of adenosine triphosphate (ATP) [14,15,16]. The propelled MTs are then able to make contact with each other to facilitate their self-assembly if suitable conditions are provided [17]. Fluorescence labeling of the MTs allows the monitoring of this assembly process by a fluorescence microscope. Thus, the in vitro gliding assay of biomolecular motors offers a self-propelled system for studying active self-assembly. In this review article, recent progress in the active self-assembly of MTs is discussed, focusing primarily on the methods developed to manipulate the interactions among the MTs and their impacts on the emergence of various types of organized structures.
Results and discussion
MTs are one of the major components of the eukaryotic cytoskeleton [5]. As MTs have the highest rigidity (2.2 × 10−23 N m2) and persistence length (~6 mm) among the cytoskeletal components, they are able to provide mechanical support to cells [18]. MTs also have a role as tracks for biomolecular motor protein-based intracellular transportation. MTs are polymers of α,β-tubulin heterodimers that join together end-to-end in a polar fashion to form protofilaments. The protofilaments of tubulins then associate laterally to form an MT, a hollow cylinder with an outer diameter of ~25 nm [5]. Although MTs in cells are composed of 13 protofilaments, the protofilament number in an MT in vitro varies depending on the polymerization conditions [19]. MTs in cells are in a dynamic equilibrium between assembly and disassembly, where one end of the MTs (plus end) grows faster than the other (minus end) [5]. On the other hand, MTs prepared in vitro can be stabilized by using a drug, Taxol, to prevent their disassembly [20]. Kinesin and dynein are two families of motor proteins that act as ATPases and interact with MTs by consuming the energy obtained from ATP hydrolysis. Kinesin-1 is the most familiar member of the kinesin family and is a dimeric, processive, plus end-directed motor protein, whereas dynein is a minus end-directed motor protein of MTs [5]. In recent years, both kinesin-1 and dynein have been employed in the active self-assembly of MTs based on the in vitro gliding assay. In the active self-assembly of MTs during the in vitro gliding assay on motor proteins, two strategies have captured attention and have been successfully applied to form various large-scale, complex and stable structures from MTs, such as bundles, rings or spools, networks, and streams through regulation of the interactions among the MTs. One method involves the functionalization of MTs with crosslinker molecules; a strong interaction between streptavidin protein and biotin was used to induce attractive interactions among the MTs gliding on a kinesin-coated substrate [21]. The other is based on the use of depletant molecules, e.g., methylcellulose (MC), which can impart a depletion force to MTs [22]. Biotin, or vitamin H, binds streptavidin with a very strong affinity (dissociation constant, Kd~10−14) and a lifetime of several hours [23]. The demonstration of the active self-assembly of MTs using the streptavidin-biotin interaction requires prior functionalization of the MTs with biotin. When biotin-functionalized MTs are allowed to glide on a kinesin motor-coated substrate, they can interact in the presence of streptavidin, ultimately resulting in active self-assembly (Fig. 1a). The initial collision of the MTs provides them with an opportunity to align and form noncovalent bonds among the colliding MTs, resulting in the formation of bundles of MTs (Fig. 1a, b). Similar to the single MTs, the MT bundles are able to exhibit translational motion, and with time they may grow in length or thickness through collision with other MTs or MT bundles. In optimized conditions, the MT bundles can be transformed into wires of millimeter-scale dimension [24]. Sometimes other structures such as MT rings or spools and network structures are reported to emerge in active self-assembly (Fig. 1b). A number of studies have been carried out to explore how to control the emergence of these various structures and to unravel the factors that control the structural polymorphism in the active self-assembly of MTs. The concentration of MTs employed in the assembly process and the ratio of streptavidin to biotin were the important factors that regulated the polymorphism of the active self-assembly [25, 26].
Among the various types of self-assembled MT structures, the rings or spools are fascinating in that they can transform the linear motion of motors into rotational motion in either a clockwise or an anticlockwise direction. The rings or spools of MTs were assembled not only on a kinesin-coated substrate but also on dyneins (Fig. 2a) [27]. The success of active self-assembly depends on the mobility of MTs on a motor protein-coated surface. In this regard, the hydrophilicity of the substrate surface appears to be an important factor. Strong hydrophobic interactions between the dynein and the cover glass surface hindered the motility of MTs, although no such difficulty was observed for kinesin. The ring-shaped MT assemblies formed on kinesin showed a preference for counterclockwise rotational motion. However, the ring-shaped MT assemblies obtained on dynein showed no preference in the direction of rotational motion, and the percentages of rings rotating in the CW and CCW directions were almost equal. This difference could be ascribed to the interaction mechanisms of kinesin and dynein with MTs. Kinesin moves along the MTs in a highly coordinated manner because of its high processivity, whereas the less processive dynein shows a highly variable stepping pattern along MTs. Typically, the size of MT rings assembled using streptavidin/biotin interaction varies over a wide range (1–12.6 μm in diameter) [26, 28]. In contrast, the formation of MT rings with a very narrow size distribution (1.8 μm with a standard deviation of 0.4 μm) has been realized at an air-buffer interface without making use of the streptavidin/biotin interaction (Fig. 2b) [29]. The formation of MT rings using the streptavidin/biotin system has been depicted by a phase diagram [25] correlating the design parameters, but there are also other reasons for MT ring formation, as reported in the literature, including the crosslinking of multiple MTs or MT bundles in closed loops and the pinning of MTs by defects in the in vitro motility assay [30, 31]. A large body of works have explored various characteristics of the MT rings, including controlling the direction of rotation, size, thickness, bias of rotation, etc. [28, 32,33,34]. The direction of rotation of the motion of MT rings is primarily governed by the helical structure of the MTs. Unlike the MTs in cells, the MTs prepared in vitro are composed of a variable number of protofilaments [19]. With the change in the protofilament number from 13, the MTs attain a left- or right-handed supertwist in their protofilament arrangement, which in turn affects their rotational direction [31]. The rotational direction and velocity of MT rings could be tuned by changing the supertwist structure of MTs by varying the preparation conditions of MTs [30]. Furthermore, the length of the MTs and the concentration and type of kinesin (as there are variations in the structure of the kinesin tail region) have a profound influence on the bias of rotation of the MT rings [32]. Furthermore, the size of the MT rings (inner diameter) is closely related to the length and rigidity of the MTs employed in the active self-assembly (Fig. 2c, d) [35]. An increase in the length and rigidity of the MTs used in self-assembly can increase the size of the MT rings (Fig. 2c), which was also supported by models and simulations [28]. In addition, incorporating MTs into the active self-assembly in a successive manner has been proven to be successful in controlling the thickness of the MT rings [32] (Fig. 2e). The MT rings are perfect examples of non-equilibrium structures with energy storage. However, many of the features of those structures, such as the distribution of their sizes [35] and the direction of rotation, are yet to be fully controlled.
a Ring-shaped MT structures produced through active self-assembly on dyneins using streptavidin-biotin interaction. Scale bar: 20 μm. Reproduced from ref. 27 with permission. b In an in vitro gliding assay on kinesins, MT filaments were transformed into ring-shaped assemblies when the filaments were subjected to an air-buffer interface. Scale bar: 25 μm. Ref. [29] reproduced by permission of The Royal Society of Chemistry. c Effect of MT length on the size of the MT rings produced through active self-assembly using streptavidin and biotin. Here, (i) to (iv) represent fluorescence microscopy images of MTs with gradually shortened lengths and (v) to (viii) represent images of MT rings for the corresponding MTs, respectively. The size of MT rings decreased with decreasing length of the MT filaments. Scale bar: 20 μm. Ref. [35] reproduced by permission of The Royal Society of Chemistry. d Average diameter of ring-shaped MT assemblies obtained from the active self-assembly of length-controlled Taxol stabilized GMPCPP-MTs (square), GMPCPP-MTs (triangle), and Taxol stabilized GTP-MTs (circle). The difference in the size of ring-shaped MT assemblies due to changes in the length and rigidity of MTs was statistically significant (*P < 0.01, #P < 0.05 using Student’s t-test). Error bar: SEM. Ref. [35] reproduced by permission of The Royal Society of Chemistry. e Fluorescence microscopy images show changes in the thickness of MT rings upon successive active self-assembly. Here, (i) to (vi) represent the first to sixth successive active self-assembly steps of MTs. Scale bar: 25 μm. Ref. [33] reproduced by permission of The Royal Society of Chemistry
We now discuss the second strategy used for the active self-assembly of MTs. Instead of using crosslinker-functionalized MTs, this approach is based on the employment of depletion force among the MTs in the in vitro gliding assay, where a high density of MTs is a prerequisite. Depletion force is an attractive interaction between colloidal particles or macromolecules suspended in a polymer solution such as MC or polyethylene glycol [36]. In this approach to active self-assembly, MTs are allowed to travel on a motor protein-coated substrate in the presence of depleting molecules, e.g., MC [22] (Fig. 3a). In these experiments, MC was used as purchased without any further treatment. At the onset of gliding motion, the MTs travel randomly without preference for any particular direction, which represents an isotropic phase (Fig. 3b). Soon, a nematic phase arises, in which the MTs self-assemble into groups much larger in size than the individual MT filaments (Fig. 3b). The structure of the MT assemblies in the nematic phase was found to be a two-dimensional structure composed of overlapping bundles of MTs [37]. In the nematic phase, the MTs exhibit preferential collective motion and over time produce various fascinating patterns, e.g., streams and vortexes [38,39,40]. Several factors, such as the density of MTs and kinesin, and the concentration of MC, have important roles in the active self-assembly of MTs in the form of collective motion [22]. To allow the emergence of collective motion, the density of MTs must be maintained above a critical value of ∼28 × 104 mm−2 [22]. MC has a crucial role in the collective motion of MTs by regulating the dynamic behavior of gliding MTs and their mutual interactions in a concentration-dependent manner. While moving on kinesins, the MTs randomly approach each other and collide, resulting in two types of events: snuggling and crossing over [22]. In the absence of MC, the probability of crossing over was much higher than that of snuggling. In a snuggling interaction, gliding MTs interact in a parallel or an antiparallel alignment. MC has a profound influence on the gliding behavior of the MTs. For example, in the presence of 0.1 wt% MC, the probability of a snuggling event was 30% and increasing the MC concentration further to 0.3 wt% increased the probability of snuggling to 50%. Snuggling is considered the most important behavior of gliding MTs for producing collective motion [38]. Thus, an increased probability of snuggling at higher MC concentrations facilitates the emergence of collective motion. The depletion force between two MTs induced by 0.3 wt% MC could be estimated from the excluded volume associated with each MTs by treating them as cylinders with a radius of ~12.5 nm surrounded by a depletion layer. The radius of gyration of 140 kDa MC is ∼30 nm. According to the Asakura–Oosawa model [41], flexible polymers such as MC are typically treated as freely interpenetrating hard spheres, which are excluded from the colloid surface by a thin layer. Then, the size of the depletion force was calculated to be ∼0.11 pN, and given the force of each kinesin ∼5 pN, the total force of kinesins interacting with each MTs on the surface should be several tens of pN. Thus, the total force of kinesin is much larger than the depletion force between the two MTs.
a Schematic representation of the active self-assembly of MTs in the presence of a depletent, methylcellulose. Ref. [22] reproduced by permission of The Royal Society of Chemistry. b Fluorescence microscopy images of MTs in an (i) isotropic and (ii) nematic phase. The nematic phase consisted of self-assembled MTs with collective motion. Scale bar: 50 μm. Ref. [22] reproduced by permission of The Royal Society of Chemistry. c Change in the orientation of MTs with time upon changing the concentration of MTs and methylcellulose. Ref. [37] published by The Royal Society of Chemistry
The kinetics of the emergence of collective motion depends on the concentrations of MTs and MC employed in the gliding assay [37]. High concentrations of MTs or MC can substantially accelerate the kinetics of the orientation process, i.e., the “isotropic to nematic” phase transition of MTs (Fig. 3c). Moreover, a minimal concentration of both MTs and MC is required to demonstrate the collective motion. The minimum concentration of MC required to induce the collective motion of MTs decreased to lower values when the concentration of MTs was very high. On the other hand, a minimum concentration of MTs was found, at or below which no collective motion could be observed even if the concentration of MC was very high. This minimum concentration of MTs was also dependent on the concentration of MC employed in the gliding assay. The effect of the concentration of MTs and MC on the kinetics of phase transition of the MTs was clearly evident from the heat maps of MT orientation at different observation times (Fig. 3c) [37]. The patterns of self-assembled MTs that emerged through the collective motions could be stabilized by using the MT-associated protein MAP4 [22]. In the presence of the 0.25 μM MAP4 fragment with 0.3 wt% MC, MTs formed streams that were stable for more than 2 h. However, in the presence of the 2.5 μM MAP4 fragment, MTs formed small bundles instead of streams and the bundles were easily detached from the surface.
In addition to the physicochemical parameters discussed above, external factors such as geometric confinement [42] or confinement by asymmetric experimental conditions [43] also have a significant effect on the active self-assembly of MTs. Geometric confinement with different shapes or sizes, fabricated using photolithographic techniques, can directly regulate the orientation of the MTs. An experimental system was developed on a micropatterned glass surface to explore the effect of confinement on the active self-assembly of MTs (Fig. 4a). The glass substrate was fabricated using photolithography to create a hydrophilic/hydrophobic pattern such that hydrophilic regions of definite shape and size were patterned on the glass substrate coated with the hydrophobic fluoropolymer, Cytop. Then, a flow cell was fabricated on the patterned glass and sequentially filled with buffers containing kinesins and MTs. Cytop reduces the nonspecific binding of proteins, allowing the attachment of MTs to the hydrophilic part of the patterned surface. A mixture of lipid-containing mineral oil was then passed through the flow cell to allow a lipid layer to cover the surface of the MT buffer droplet due to the amphiphilic nature of the lipid. As a result, a semispherical droplet of MT buffers was confined on the hydrophilic region of the patterned glass surface covered by the lipid layer, making a confined space. All experiments were repeated several times, and the reproducibility of the results was confirmed. The self-assembled MTs were found to align preferentially in particular directions under geometrical confinement. The aligned MTs demonstrated different orientations inside confined areas of different shapes and sizes (Fig. 4b, c). In circular confined areas, MTs were preferentially aligned either along the periphery or at the center of the circle. When confined in a square, triangle, rectangle with sharp edge, hexagon, rectangle with round edge or star, the MTs showed two types of organization behaviors, as observed in the circle. MTs were aligned along the wall around the periphery and along the diagonal around the center of the shapes. Analysis of the orientation angles revealed that the MTs showed similar types of orientation in the square, triangle, rectangle with sharp edges and hexagons. The MTs were oriented mostly in two directions. At the center, the MTs were aligned along the diagonal of the confined area and oriented in a particular direction. On the other hand, the orientation of MTs in the rectangle with a round edge was almost the same as that observed in the circular shape. In the star-shaped confined area, the MTs were found to orient along the four vertices of the star. The distinct orientations of MTs could be accounted for by the differences in the geometry of the confined areas in different shapes. In addition to the shape, the size of the confined area also greatly influences the self-assembly and orientation of MTs (Fig. 4c).
a Schematic diagram of the experimental design employed to demonstrate the active self-assembly of MTs in a confined space. Ref. [42] reproduced with permission. b Fluorescence microscopy images of self-assembled MTs inside confined areas with the circle, square, triangle, rectangle with sharp edge, hexagon, rectangle with round edge and star-shapedareas. Shape size: 200 μm. Ref. [42] adapted with permission. c Effect of the size of the confined area on the active self-assembly of MTs. MTs were allowed to move inside circular confined areas of varying size. Scale bar: 10 μm. Ref. [42] reproduced with permission. d Schematic diagram of the experimental design used to demonstrate the active self-assembly of microtubules at an air-buffer interface. Ref. [43] reproduced by permission of The Royal Society of Chemistry. e Fluorescence microscopy image of MT vortices produced at the air-buffer interface. Ref. [43] reproduced by permission of The Royal Society of Chemistry. Scale bar: 2.5 μm. f Time-dependent change in MT vortices at the air-buffer interface. Ref. [43] reproduced by permission of The Royal Society of Chemistry. Scale bar: 2.5 μm
On the other hand, confinement under asymmetric assembly conditions facilitated the morphological transition of MTs into rings or vortices. MT rings with very small circumferences were formed when MT filaments were subjected to an incompatible environment, e.g., an air-buffer interface [43] (Fig. 4d, e). The required driving force for this morphological transformation of MTs could be explained by the need for MT protein filaments to reduce their energy in an incompatible environmental condition (air). Interestingly, these MT rings exhibited dynamic transformation with time (Fig. 4f), following four different routes [43]. The most striking feature of the MT rings formed at the air-buffer interface is that they demonstrated periodic mechanical oscillation, which is ascribed to the mechanical frustration of the dynamic rings. Crowding effects and interactions of MTs with their nearest neighbors in a high-density condition can also act as the driving force in the formation of MT vortices [38, 40].
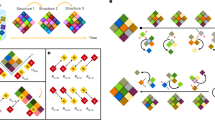
Reversibility is a salient feature of self-assembly in nature that can be observed in the swarming of fish schools, ant colonies, bird flocks, and so on [44,45,46]. The demonstration of self-assembly in such a reversible fashion requires the ability to control the local interactions among the building blocks, which is a challenging task in fabrication-based conventional self-assembly or supramolecular chemistry. In a recent study, programmability was successfully introduced into the dynamic self-assembly of MTs [47]. DNA was employed as a universal interface to reversibly regulate the self-assembly of MTs in a programmable manner. DNA is a storage device for genetic information and has emerged as a versatile tool for molecular computing. Recent advances in chemical DNA synthesis have rendered DNA a potential building block for the precise construction of nanostructures [48, 49]. By tethering single-strand DNA to MTs as an information processor, it has been possible to control the active self-assembly of MTs by programming their interactions. Here, the active self-assembly of MTs was demonstrated based on the in vitro gliding assay of MTs as discussed above. However, the MTs were conjugated with two single-stranded DNAs, termed “receptor DNA” (r-DNA), of prescribed base number, e.g., T16 and (TTG)5, through a copper-free click reaction. The DNA-conjugated MTs were propelled similarly to the bare MTs by surface-adhered kinesins, using the chemical energy of ATP, without sacrificing their velocity. Kinesin is an ATPase, i.e., it can consume ATP and release ADP and inorganic phosphate. Kinesins can move towards the plus end of MTs by converting the chemical energy of ATP hydrolysis into mechanical work [5, 18]. When adsorbed to a surface, kinesin can generate force (~5 pN) by consuming ATP, and thus MTs are made to move on the kinesins in the in vitro gliding assay. Dyneins are also ATPase- and MT-associated motor proteins, but unlike kinesins, dyneins move towards the minus end of MTs. Therefore, on a kinesin-coated surface, gliding MTs move with their minus end as the leading end, whereas the plus end of MTs becomes the leading end when MTs are made to move on a dynein-coated substrate. Linker DNA (l-DNA) was designed to be partially complementary to the r-DNAs and able to crosslink them through hybridization (Fig. 5a). The self-assembly of the gliding MTs was initiated by the introduction of l-DNA. While gliding, the r-DNA-conjugated MTs collided and formed bundles (Fig. 5b), and the size of the bundles increased over time. Despite the increase in size, the MT bundles exhibited translational motion with a velocity (0.51 ± 0.02 µm s−1) close to that of individual MTs (0.60 ± 0.05 µm s−1). Ring-shaped MT assemblies were produced through repeated collision of the bundles (Fig. 5b). From the count of the number of individual MTs at different times, the association ratio was calculated as the fraction of the number of MTs initially incorporated into the swarms. The active self-assembly was dependent on the concentration of r-DNAs, l-DNA, and the density of MTs, suggesting that all these components play significant roles in the self-assembly of the DNA-conjugated MTs. Reversibility in the self-assembly of MTs was demonstrated by introducing a dissociation DNA (d-DNA), which was designed to extract the l-DNA through a DNA strand exchange reaction. Within a few minutes after the addition of the d-DNA, the self-assembled MT rings were dissociated into isolated MTs upon (Fig. 5c). The utility of DNA as an operator for molecular computing further allowed the demonstration of different logical operations, e.g., YES, AND, and OR logic gates, through the active self-assembly of MTs. Moreover, by precisely tuning the physical properties of MTs, a reversible bundle-shaped assembly of MTs was obtained. The incorporation of the photoresponsive molecule azobenzene into the DNA facilitated reversible and noninvasive control of the active self-assembly of MTs for multiple cycles by photoirradiation (Fig. 6a). This photoregulation of the dynamic self-assembly of MTs was successful for both MT rings and bundles (Fig. 6b, c).
a Schematic representation of the experimental design employed to demonstrate the DNA-regulated active self-assembly and disassembly of microtubules. Adapted from ref. [47] with permission. b Formation of ring-shaped MT assemblies with time using DNA-based interactions. Adapted from ref. [47] with permission. c Disassembly of an MT ring with time using dissociation DNA, which mediates the DNA strand displacement reaction. Adapted from ref. [47] with permission. Scale bar: 20 μm
a Reversible hydrogen bonding between photoresponsive DNAs by the light-induced cis–trans isomerization of azobenzene. Adapted from ref. [47] with permission. Photoregulated repeated assembly and disassembly of b microtubule bundles and c rings. Visible light induced the assembly of rigid and flexible MT filaments into bundles and rings, respectively. Upon UV light irradiation, the bundles and rings of MTs were disassembled into the filament MTs. Adapted from ref. [47] with permission. Scale bar: 20 μm
Summary and outlook
In this review article, the latest advances in the active self-assembly of MTs have been summarized. Various methods developed in recent years for demonstrating the active self-assembly of the self-propelled bimolecular motor system have been comprehensively discussed. The methods are broadly categorized into three main groups depending on the mechanisms through which interactions among the MTs are regulated in the assembly process. First, the active self-assembly of MTs by employing a strong interaction between streptavidin and biotin was discussed. This method has provided self-assembled structures with rich morphological variations, i.e., bundles, rings, and networks. In another approach, utilizing depletion force, a large number of MTs were self-assembled into gigantic stream or vortex patterns. However, both of these methods have limitations in that they were unable to allow reversibility in the assembly processes, as nature does. In the latest approach, a solution to this limitation was provided. Utilizing the potential of DNA for logical operations, molecular computations and high selectivity, reversible regulation of the active self-assembly of MTs in a programmable manner was realized using both chemical and physical input signals. This advance should impact self-assembly not only in the field of biomolecular motors but also in supramolecular chemistry and in bio- and DNA-related nanotechnology. The application of biomolecular motors in active self-assembly has greatly benefited our understanding of self-assembly in nature. Although we now have greatly improved control over the characteristics of the structures created through self-assembly, the variety of the structures is still quite limited compared to that available in nature. Further exploration is required to address the need for more precise control of the emergence of the structures and their prospective applications in an artificial environment.
References
Kushner DJ. Self-assembly of biological structures. Bacteriol Rev. 1969;33:302–45.
Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295:2418–21.
Fialkowski M, Bishop KJ, Klajn R, Smoukov SK, Campbell CJ, Grzybowski BA. Principles and implementations of dissipative (dynamic) self-assembly. J Phys Chem B. 2006;110:2482–496.
Hess H. Self-assembly driven by molecular motors. Soft Matter. 2006;2:669–77.
Alberts, B et al. Molecular biology of the cell, Garland Science, New York, 2008.
Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400.
Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol. 2017;18:187–201.
Knoblauch M, Peters WS. Biomimetic actuators: where technology and cell biology merge. Cell Mol Life Sci. 2004;61:24972509.
Hess H. Engineering applications of biomolecular motors. Annu Rev Biomed Eng. 2011;13:429.
Lam AT, VanDelinder V, Kabir AMR, Hess H, Bachand GD, Kakugo A. Soft Matter. 2016;12:988.
Hess H, Ross JL. Non-equilibrium assembly of microtubules: from molecules to autonomous chemical robots. Chem Soc Rev. 2017;46:5570–87.
Ndlec FJ, Surrey T, Maggs AC, Leibler S. Self-organization of microtubules and motors. Nature. 1997;389:305–8.
Kabir AMR, Kakugo A, Gong JP, Osada Y. How to integrate biological motors towards bio-actuators fueled by ATP. Macromol Biosci. 2011;11:1314–24.
Kron SJ, Toyoshima YY, Uyeda TQ, Spudich JA. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 1991;196:399–416.
Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci USA. 1986;83:6272–6.
Kabir AMR, Inoue D, Kakugo A, Kamei A, Gong JP. Prolongation of the active lifetime of a biomolecular motor for in vitro motility assay by using an inert atmosphere. Langmuir. 2011;27:13659–68.
Kabir AMR, Inoue D, Kakugo A, Sada K, Gong JP. Active self-organization of microtubules in an inert chamber system. Polym J. 2012;44:607–11.
Howard, J. Mechanics of motor proteins and the cytoskeleton, Sinauer Associates Inc., Sunderland, 2001.
Pierson GB, Burton PR, Himes RH. Alterations in number of protofilaments in microtubules assembled in vitro. J Cell Biol. 1978;76:223–8.
Arnal I, Wade RH. How does taxol stabilize microtubules? Curr Biol. 1995;5:900–8.
Hess H, Clemmens J, Brunner C, Doot R, Luna S, Ernst KH, et al. Molecular self-assembly of “nanowires” and “nanospools” using active transport. Nano Lett. 2005;5:629–33.
Inoue D, Mahmot B, Kabir AMR, Farhana TI, Tokuraku K, Sada K, et al. Depletion force induced collective motion of microtubules driven by kinesin. Nanoscale. 2015;7:18054–61.
He S, Lam AT, Jeune-Smith Y, Hess H. Modeling negative cooperativity in streptavidin adsorption onto biotinylated microtubules. Langmuir. 2012;28:10635–10639.
Idan O, Lam A, Kamcev J, Gonzales J, Agarwal A, Hess H. Nanoscale transport enables active self-assembly of millimeter-scale wires. Nano Lett. 2011;12:240–5.
Tamura Y, Kawamura R, Shikinaka K, Kakugo A, Osada Y, Gong JP, et al. Dynamic self-organization and polymorphism of microtubule assembly through active interactions with kinesin. Soft Matter. 2011;7:5654–9.
Kawamura R, Kakugo A, Osada Y, Gong JP. Microtubule bundle formation driven by ATP: the effect of concentrations of kinesin, streptavidin and microtubules. Nanotechnology. 2010;21:145603.
Ito M, Kabir AMR, Inoue D, Torisawa T, Toyoshima Y, Sada K, et al. Formation of ring-shaped microtubule assemblies through active self-organization on dynein. Polym J. 2014;46:220–5.
Kawamura R, Kakugo A, Shikinaka K, Osada Y, Gong JP. Ring-shaped assembly of microtubules shows preferential counterclockwise motion. Biomacromolecules. 2008;9:2277–82.
Kabir AMR, Wada S, Inoue D, Tamura Y, Kajihara T, Mayama H, et al. Formation of ring-shaped assembly of microtubules with a narrow size distribution at an air-buffer interface. Soft Matter. 2012;8:10863–7.
Luria I, Crenshaw J, Downs M, Agarwal A, Seshadri SB, Gonzales J, et al. Microtubule nanospool formation by active self-assembly is not initiated by thermal activation. Soft Matter. 2011;7:3108–15.
Lam AT, Curschellas C, Krovvidi D, Hess H. Controlling self-assembly of microtubule spools via kinesin motor density. Soft Matter. 2014;10:8731–6.
Kakugo A, Kabir AMR, Hosoda N, Shikinaka K, Gong JP. Controlled clockwise-counterclockwise motion of the ring-shaped microtubules assembly. Biomacromolecules. 2011;12:3394–9.
Inoue D, Kabir AMR, Mayama H, Gong JP, Sada K, Kakugo A. Growth of ring-shaped microtubule assemblies through stepwise active self-organization. Soft Matter. 2013;9:7061–8.
Wada S, Kabir AMR, Kawamura R, Ito M, Inoue D, Sada K, et al. Controlling the bias of rotational motion of ring-shaped microtubule assembly. Biomacromolecules. 2014;16:374–8.
Wada S, Kabir AMR, Ito M, Inoue D, Sada K, Kakugo A. Effect of length and rigidity of microtubules on the size of ring-shaped assemblies obtained through active self-organization. Soft Matter. 2015;11:1151–7.
Mao Y, Cates ME, Lekkerkerker HNW. Depletion force in colloidal systems. Phys A: Stat Mech its Appl. 1995;222:10–24.
Saito A, Farhana TI, Kabir AMR, Inoue D, Konagaya A, Sada K, et al. Understanding the emergence of collective motion of microtubules driven by kinesins: role of concentration of microtubules and depletion force, 2017. RSC Adv. 2017;7:13191–7.
Sumino Y, Nagai KH, Shitaka Y, Tanaka D, Yoshikawa K, Chate H, et al. Large-scale vortex lattice emerging from collectively moving microtubules. Nature. 2012;483:448–52.
Liu L, Tuzel E, Ross JL. Loop formation of microtubules during gliding at high density. J Phys Condens Matter. 2011;23:374104.
Schaller V, Weber C, Semmrich C, Frey E, Bausch AR. Polar patterns of driven filaments. Nature. 2010;467:73–77.
Asakura S, Oosawa F. On interaction between two bodies immersed in a solution of macromolecules. J Chem Phys. 1954;22:1255–6.
Islam MS, Kuribayashi-Shigetomi K, Kabir AMR, Inoue D, Sada K, Kakugo A. Role of confinement in the active self-organization of kinesin-driven microtubules. Sens Actuators B Chem. 2017;247:53–60.
Ito M, Kabir AMR, Islam MS, Inoue D, Wada S, Sada K, et al. Mechanical oscillation of dynamic microtubule rings. RSC Adv. 2016;6:69149–55.
Bonabeau E, Dorigo M, Theraulaz G. Swarm Intelligence: From Natural to Artificial Systems. Oxford, New York: Oxford Univ. Press; 1999.
Beshers SN, Fewell JH. Models of division of labor in social insects. Annu Rev Entomol. 2001;46:413–40.
Niven JE. How honeybees break a decision-making deadlock. Science. 2011;335:43–44.
Keya JJ, Suzuki R, Kabir AMR, Inoue D, Asanuma H, Sada K, et al. DNA-assisted swarm control in a biomolecular motor system. Nat Commun. 2018;9:4531–8
Seeman NC. Nucleic-acid junctions and lattices. J Theor Biol. 1982;99:237–47.
Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302.
Acknowledgements
This work was financially supported by a Grant-in-Aid for Scientific Research on Innovative Areas “Molecular Robotics” (JSPS KAKENHI Grant Number JP24104004), a Grant-in-Aid for Challenging Exploratory Research (JSPS KAKENHI Grant Number JP15K12135) and a Grant-in-Aid for Scientific Research (B) (JSPS KAKENHI Grant Number JP15H03706) from Japan Society for the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kabir, A.M.R., Kakugo, A. Study of active self-assembly using biomolecular motors. Polym J 50, 1139–1148 (2018). https://doi.org/10.1038/s41428-018-0109-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41428-018-0109-8
This article is cited by
-
Cyclic Tau-derived peptides for stabilization of microtubules
Polymer Journal (2020)