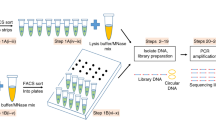
Abstract
Although three-dimensional (3D) genome organization is central to many aspects of nuclear function, it has been difficult to measure at the single-cell level. To address this, we developed ‘single-cell split-pool recognition of interactions by tag extension’ (scSPRITE). scSPRITE uses split-and-pool barcoding to tag DNA fragments in the same nucleus and their 3D spatial arrangement. Because scSPRITE measures multiway DNA contacts, it generates higher-resolution maps within an individual cell than can be achieved by proximity ligation. We applied scSPRITE to thousands of mouse embryonic stem cells and detected known genome structures, including chromosome territories, active and inactive compartments, and topologically associating domains (TADs) as well as long-range inter-chromosomal structures organized around various nuclear bodies. We observe that these structures exhibit different levels of heterogeneity across the population, with TADs representing dynamic units of genome organization across cells. We expect that scSPRITE will be a critical tool for studying genome structure within heterogeneous populations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets (Figs. 1–5 and Extended Data Figs. 1−5) generated and analyzed in the current study are available in the Gene Expression Omnibus repository under accession number GSE154353 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154353).
Code availability
scSPRITE software is available at https://github.com/caltech-bioinformatics-resource-center/Guttman_Ismagilov_Labs.
References
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289 (2009).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Dekker, J. & Mirny, L. The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 (2016).
Freire-Pritchett, P. et al. Global reorganisation of cis-regulatory units upon lineage commitment of human embryonic stem cells. eLife 6, e21926 (2017).
Whyte, WarrenA. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Quinodoz, S. A. et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e724 (2018).
Mao, Y. S., Zhang, B. & Spector, D. L. Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306 (2011).
Miura, H. et al. Single-cell DNA replication profiling identifies spatiotemporal developmental dynamics of chromosome organization. Nat. Genet. 51, 1356–1368 (2019).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Chen, Y. et al. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 217, 4025–4048 (2018).
Pederson, T. The nucleolus. Cold Spring Harb. Perspect. Biol. 3, a000638 (2011).
Finn, E. H. et al. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515 (2019).
Wang, S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598 (2016).
Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Nagano, T. et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017).
Ma, X., Ezer, D., Adryan, B. & Stevens, T. J. Canonical and single-cell Hi-C reveal distinct chromatin interaction sub-networks of mammalian transcription factors. Genome Biol. 19, 174 (2018).
Mohammed, H. et al. Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep. 20, 1215–1228 (2017).
Buettner, F. et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 33, 155–160 (2015).
Bintu, B. et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018).
Giorgetti, L. et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157, 950–963 (2014).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
O’Sullivan, J. M., Hendy, M. D., Pichugina, T., Wake, G. C. & Langowski, J. The statistical-mechanics of chromosome conformation capture. Nucleus 4, 390–398 (2013).
Kolodziejczyk, A. A. et al. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17, 471–485 (2015).
Guo, F. et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 27, 967–988 (2017).
Ghimire, S. et al. Comparative analysis of naive, primed and ground state pluripotency in mouse embryonic stem cells originating from the same genetic background. Sci. Rep. 8, 5884 (2018).
Lee, D.-S. et al. Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nat. Methods 16, 999–1006 (2019).
Zhou, S., Jiang, W., Zhao, Y. & Zhou, D.-X. Single-cell three-dimensional genome structures of rice gametes and unicellular zygotes. Nat. Plants 5, 795–800 (2019).
Ramani, V. et al. Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017).
Ramani, V. et al. Sci-Hi-C: a single-cell Hi-C method for mapping 3D genome organization in large number of single cells. Methods 170, 61–68 (2020).
Spector, D. L. & Lamond, A. I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646 (2011).
Guenatri, M., Bailly, D., Maison, C. L. & Almouzni, G. V. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166, 493–505 (2004).
Almouzni, G. & Probst, A. V. Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus 2, 332–338 (2011).
Strongin, D. E., Groudine, M. & Politz, J. C. R. Nucleolar tethering mediates pairing between the IgH and Myc loci. Nucleus 5, 474–481 (2014).
Dowen, J. M. et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387 (2014).
Pachano, T., Crispatzu, G. & Rada-Iglesias, A. Polycomb proteins as organizers of 3D genome architecture in embryonic stem cells. Brief. Funct. Genomics 18, 358–366 (2019).
Blinka, S., Reimer, Michael, H. Jr., Pulakanti, K. & Rao, S. Super-enhancers at the Nanog locus differentially regulate neighboring pluripotency-associated genes. Cell Rep. 17, 19–28 (2016).
Novo, C. L. et al. Long-range enhancer interactions are prevalent in mouse embryonic stem cells and are reorganized upon pluripotent state transition. Cell Rep. 22, 2615–2627 (2018).
Schoenfelder, S. et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 25, 582–597 (2015).
Apostolou, E. et al. Genome-wide chromatin interactions of the nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell 12, 699–712 (2013).
Russell, R. et al. A dynamic role of TBX3 in the pluripotency circuitry. Stem Cell Rep. 5, 1155–1170 (2015).
Kalmar, T. et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149 (2009).
Engreitz, J. M. et al. RNA–RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell 159, 188–199 (2014).
Li, W., Gong, K., Li, Q., Alber, F. & Zhou, X. J. Hi-Corrector: a fast, scalable and memory-efficient package for normalizing large-scale Hi-C data. Bioinformatics 31, 960–962 (2015).
Acknowledgements
We would like to thank F. Gao from Caltech’s Bioinformatics Resource Center and I. Antoshechkin from Caltech’s Millard and Muriel Jacobs Genetics and Genomics Laboratory for assistance. We would also like to thank C. Chen, V. Trinh, E. Detmar, E. Soehalim, A. Narayanan and I. Goronzy for their contributions in helping develop scSPRITE and analysis. We would like to thank M. Thompson’s laboratory for allowing us to use their MiSeq instrument and the ENCODE Consortium and the ENCODE production laboratory of B. Ren (University of California, San Diego) for making their data publicly available. We also thank N. Shelby and S. Hiley for contributions to the writing and editing this manuscript and I.-M. Strazhnik for helping with illustrations. Funding: This work was funded by the National Institutes of Health 4DN Program (U01 DA040612 and U01 HL130007), the National Human Genome Research Institute Genomics of Gene Regulation Program (U01 HG007910), the New York Stem Cell Foundation (NYSCF-R-I13), the Sontag Foundation and funds from Caltech. M.V.A. and S.A.Q. were funded by a National Science Foundation Graduate Research Fellowship Program fellowship. M.V.A. was additionally funded by the Earle C. Anthony Fellowship (Caltech). M. Guttman is an NYSCF-Robertson Investigator.
Author information
Authors and Affiliations
Contributions
M.V.A. conducted the experiments to develop and validate the method, conceptualized and performed the analyses and wrote the manuscript. J.W.J. contributed to and supervised the experiments to develop and validate the method, conceptualized and performed the analyses and wrote the manuscript. N.O. conceptualized and performed analysis to validate the method, developed the pipeline for the workup of scSPRITE sequencing data and contributed to writing the manuscript. M.S.C. contributed to the experiments to develop the method. C.A.L. developed a pipeline to sort cells by cell-specific barcodes. S.A.Q. contributed to the experiments to develop and validate the method. D.A.S. contributed to conceptualize scSPRITE and to the experiments to develop the method. R.F.I. conceptualized scSPRITE and supervised the experiments and the analysis to develop the method. M.G. conceptualized scSPRITE, supervised the experiments and the analysis to validate the method and wrote the manuscript. For detailed author contributions, please see Supplementary Note 5.
Corresponding authors
Ethics declarations
Competing interests
This paper is the subject of a patent application filed by Caltech. R.F.I. has a financial interest in Talis Biomedical Corp. S.A.Q. and M.G. are inventors on a patent owned by Caltech on SPRITE. The remaining authors declare no competing financial interests.
Additional information
Peer review information Nature Biotechnology thanks Andrew Adey and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 scSPRITE generate single cell maps with high genomic coverage.
a. Quantification of cell aggregation. Top: number of cells in clumps pre- and post-filtration (singlets, doublets, triplets, etc). Bottom: microscope images (10x) of cells pre- and post-filtration step, scale bar 100 µm. b. Validation of In-nuclei barcoding step of the protocol on mixed cell population (human-mouse cells): no mixing (top middle and top right), mixing before crosslinking (bottom left), mixing after crosslinking (bottom middle), and mixing after in-nuclei restriction digest (bottom right). c. Schematic of the computational analysis pipeline for processing scSPRITE data. d. Theoretical number of contacts measured by SPRITE-derived methods and Hi-C-derived methods over increasing numbers of DNA molecules per complex. e. Maximum number of pairwise interactions that can be obtained from proximity ligation (Hi-C-derived methods) and complex barcoding (SPRITE-derived methods). f. Genome-wide coverage for the filtered 1,000 cells: the median (black triangular points) and median absolute deviation (MAD) (green circular points) values were calculated per cell using the number of reads per 1 Mb bin genome-wide (chr1-19). g. Genomic coverage of 20 random cell barcodes; 1 Mb bin per chromosome.
Extended Data Fig. 2 Known chromosomal structures can be measured genome-wide in hundreds of single mESCs by scSPRITE.
a. Additional single cell examples of chromosome territory structure between chr1 and chr2; plotted as number of DNA clusters at 1 Mb resolution. Box plot represents normalized detection scores between chr1 and chr2, where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). b. Chromosome territory scores across 1000 cells (clustered based on similarity pattern). Columns represent chromosome territory detection scores for all pairs of chromosomes with the reference chromosome. Arrows represent chromosome territory scores between chr1 and chr2, which were analyzed in this paper. c. Quantification of chromosome territory scores with respect to each chromosome. Boxplots show the range of chromosome territory scores, the average score (black line), and individual pairs of chromosome territory scores (grey dots). d. Box plot represents average chromosome territory detection scores from all genome-wide (chr1-19) chromosome pairs., where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells) (left).. Additional single cell examples of genome-wide (chr1-19) chromosome territories (right). e. Additional single cell examples of A/B compartments detected within 0-55Mb in chr2; plotted number of DNA clusters at 1 Mb resolution (right). Box plot represents normalized detection scores between 0-55Mb in chr2, where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). f. Representation of compartment switching scores across 1,000 cells (clustered based on score similarity pattern). Columns represent the strength of compartment switching detection scores for compartments that switched from “B-to-A-to-B” or “A-to-B-to-A” genome-wide (chr1-19). Arrows represent compartment switching scores for chr2 1-55 Mb, chr8 22-37 Mb, chr10 58-70 Mb, and chr17 8-45 Mb, all of which were analyzed in this paper. g. Additional single cell examples of compartment switching from Region 1, Region 2, and Region 3 (right). For each region’s box plot: whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). h. Expected (right) and observed (left) coverage of reads in the A and B compartment.
Extended Data Fig. 3 Higher-order structures are identified genome-wide in hundreds of single mESC by scSPRITE method.
a. Additional single cell examples of nucleolar interactions detected between chr18 and chr19; plotted number of DNA clusters at 1 Mb resolution; detection scores below contact map (right). Box plot represents normalized detection scores between chr18 and chr19, where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). b. Nucleolar interaction between chr12 and chr19: detection scores for 1000 cells (middle). Box plot where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). Representation of structures with max score (+1) and min. score (-1) (left) and ensemble scSPRITE heatmap (middle); contact map at 1 Mb resolution. Single cell examples (right); plotted number of DNA clusters at 1 Mb resolution. c. Relative correlation of the percent of cells from scSPRITE vs DNA-FISH containing inter-chromosomal interactions at specified 1 Mb regions targeted by DNA-FISH probes. Control chromosomes (grey points) and nucleolar associating chromosomes (black dots) are plotted. d. Relative correlation of the contact frequency from scSPRITE vs the contact frequency from SPRITE containing inter-chromosomal interactions targeted by DNA-FISH probes. Control chromosomes (grey points) and nucleolar associating chromosomes (black dots) are plotted. e. Frequency of cells containing inter-chromosomal nucleolar contacts (normalized to number of reads per region) for each pair of nucleolar associating chromosomes.. f. Single cell examples of speckle interaction detected between chr2 and chr5; plotted number of DNA clusters at 1 Mb resolution. Box plot represents normalized detection scores between chr2 and chr5, where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). g. Additional single cell examples of speckle interactions detected between chr2 and chr4; plotted number of DNA clusters at 1 Mb resolution. Box plot represents normalized detection scores between chr2 and chr4, where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). h. Frequency of cells containing inter-chromosomal speckle contacts (normalized to number of reads per region) for each pair of speckle associating chromosomes. i. Additional single cell examples of centromere-proximal interactions detected between chr1 and chr11; plotted number of DNA clusters at 1 Mb resolution. Box plot represents normalized detection scores between chr1 and chr11, where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). j. Single cell examples of chr4 and chr11 centromere-proximal regions interacting together; plotted number of DNA clusters at 1 Mb resolution. Box plot represents normalized detection scores between chr4 and chr11, where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). k. Frequency of cells containing inter-chromosomal centromeric contacts (normalized to number of reads per region) for each pair of chromosomes. l. Higher-order structures representation from scHi-C data16 – centromere-proximal interactions, speckle interactions, and nucleolar interactions; Pairwise contact map from ensemble 1,000 cells (left), pairwise contact map from their best single cell (right).
Extended Data Fig. 4 TADs are heterogeneous units present in the genomes of individual mESCs.
a. Genome-wide correlation of insulation scores between ensemble scSPRITE and Hi-C3 from mouse ES cells at 40 kb resolution. b. Insulation score profile of ensemble scSPRITE (red) and Hi-C3 (blue) at 40 kb resolution at chr1 65-95 Mb. c. Additional single cell examples of TAD-like structures between 124.8-126.7Mb of chr4; plotted number of DNA clusters at 40 kb resolution; detection scores below contact map. Box plot represents normalized detection scores between 124.8-126.7Mb of chr4, where whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median, red dots represent single cell examples (n = 1000 cells). d. TAD detection scores across 1,000 cells (clustered based on score similarity pattern) in chr2 (left) and chr18 (right). Columns represent the strength of TAD detection scores for all TADs detected across chr2 or chr18, respectively, in ensemble scSPRITE. e. TAD detection scores across 1,000 cells between 38.5-48.56 Mb of chr4. Each line represents the strength of TAD detection scores in this given region from a single cell. Cells are either in Group 1 or 2 in Fig. 4f or not used. f. Ensemble heatmap from all 1000 cells between 39.4-41.4Mb of chr4 representing strong TADs detected in bulk (blue lines), and weak emerging TADs (green line) over the A/B boundary. g. Fraction of cells in each cell cycle phase from the set of single cells containing (left) or lacking (right) the contact between the boundary region (Fig. 4f). h. Difference contact map across a control region 84.8-88.4 Mb of chr4 made by subtracting the normalized contacts from cells in Group II from Group I (Fig. 4f). Insulation scores for cells in Group I (dark grey) and Group II (light grey) are plotted.
Extended Data Fig. 5 Structural heterogeneity in long-range interactions is revealed by scSPRITE.
a. Ensemble heatmaps across 122.2-122.8 Mb region in chr6 representing cells containing (top) or lacking (bottom) the contact between the Nanog locus and the -300 Kb SE. Blue square shows the contact. b. Number of genome-wide reads (left) and number of genome-wide contacts (right) for groups of cells with and without the Nanog-SE interaction. For each box plot, whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median (with = 159 cells, without = 149 cells). No statistical significance between the two groups were seen based on the Kolmogorov–Smirnov two-sided test. c. Fraction of cells in each cell cycle phase from the set of single cells containing (left) or lacking (right) the contact between the Nanog locus and the SE 300kb upstream of Nanog. d. Heatmaps between 119.24-121.28Mb in chr5 of pooled cells either containing (top) or lacking (bottom) the contact between the Tbx3 locus and Lhx5. Blue square shows the contact. e. Number of genome-wide reads (left) and number of genome-wide contacts (right) for groups of cells with and without the Tbx3-Lhx5 interaction. For each box plot, whiskers represent the 10th and 90th percentiles, box limits represent the 25th and 75th percentiles, black line represents the median (with = 152 cells, without = 149 cells). No statistical significance between the two groups were seen based on the Kolmogorov–Smirnov two-sided test. f. Fraction of cells in each cell cycle phase from the set of single cells containing (left) or lacking (right) the contact between the Tbx3 locus and the Lhx5.
Supplementary information
Supplementary Information
Supplementary Notes 1−5
Supplementary Tables
Table 1: Metrics for each pair of chromosome territory normalized detection scores. Table 2: Metrics for each region of A/B compartment normalized detection scores. Table 3: Metrics for each TAD normalized detection score.
Rights and permissions
About this article
Cite this article
Arrastia, M.V., Jachowicz, J.W., Ollikainen, N. et al. Single-cell measurement of higher-order 3D genome organization with scSPRITE. Nat Biotechnol 40, 64–73 (2022). https://doi.org/10.1038/s41587-021-00998-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-021-00998-1
This article is cited by
-
Spatial multi-omics: novel tools to study the complexity of cardiovascular diseases
Genome Medicine (2024)
-
Combinatorial single-cell profiling of major chromatin types with MAbID
Nature Methods (2024)
-
Single-cell multiplex chromatin and RNA interactions in ageing human brain
Nature (2024)
-
Enhancer selectivity in space and time: from enhancer–promoter interactions to promoter activation
Nature Reviews Molecular Cell Biology (2024)
-
Computational methods for analysing multiscale 3D genome organization
Nature Reviews Genetics (2024)