Abstract
Generation of stable gene-edited plant lines using clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein 9 (Cas9) requires a lengthy process of outcrossing to eliminate CRISPR–Cas9-associated sequences and produce transgene-free lines. We have addressed this issue by designing fusions of Cas9 and guide RNA transcripts to tRNA-like sequence motifs that move RNAs from transgenic rootstocks to grafted wild-type shoots (scions) and achieve heritable gene editing, as demonstrated in wild-type Arabidopsis thaliana and Brassica rapa. The graft-mobile gene editing system enables the production of transgene-free offspring in one generation without the need for transgene elimination, culture recovery and selection, or use of viral editing vectors. We anticipate that using graft-mobile editing systems for transgene-free plant production may be applied to a wide range of breeding programs and crop plants.
Similar content being viewed by others
Main
Programmed by guide RNA (gRNA) sequences, the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) nuclease creates DNA double-strand breaks at specific genomic sequences, resulting in edited sequences1,2. In plants, to obtain transgene-free mutants and to stabilize the genomes, both Cas9 enzyme and gRNA must be either introduced transiently or outcrossed from a mutant progeny3. For example, functional gRNA and Cas9 complexes can be ectopically introduced into protoplasted cells or embryos from which transgene-free edited plants have to be regenerated and selected. These approaches require time-consuming plant regeneration and selection steps, or expensive reagents and special equipment4. Given that most plant species are not easily accessible to transformation or have long generation times we sought to design Cas9 and gRNA transcripts that are transported from transgenic roots to wild-type shoots (scions). On such grafted wild-type scions, gene-edited (that is, mutant) seeds might be created by gRNA and Cas9 transcripts transported from a transgenic rootstock. Transcript mobility can be introduced by tRNA-like sequence (TLS) motifs and variants thereof shown to license transport of protein encoding transcripts over graft junctions5 or to parasitic Cuscuta plants feeding on host plants6. We postulated that the addition of TLS motifs to Cas9 and gRNA transcripts will result in their root-to-shoot movement and cause editing in recipient-grafted wild-type tissues. Consequently, transgene-free scions grafted on transgenic mobile Cas9/gRNA expressing rootstocks should produce seeds with edited genomes.
Results
To test whether Cas9 transcript and gRNA can be delivered from donor rootstocks to wild-type scions (Fig. 1a), we created two gRNAs introducing a genomic deletion in the NITRATE REDUCTASE1 (NIA1, AT1G77760) gene. NIA1 is one of two Arabidopsis nitrate reductase enzymes converting nitrate (NO3) to ammonium (NH4) and on NH4-deficient medium nia1 mutants develop a clearly visible phenotype with smaller plants that become chlorotic over time7,8, which should allow us to identify nia1 mutants in progeny plants by phenotype. We generated Arabidopsis thaliana lines expressing zCas99 from an estradiol-inducible promoter and two NIA1-targeting gRNAs (gNIA1) driven by the constitutive Pol-III promoters U6-26 and U6-2910. The two NIA1-targeting gRNAs are designed to create double genome edits (deletions) resulting in the deletion of approximately 1,000 base pairs (bp) of the NIA1 gene sequence creating a nia1 knock out. Additionally, we constructed gRNAs targeting a transgenic 35Spromoter::Venus::35Sterminator::BASTA construct. The two gRNAs are designed to introduce a deletion removing the Venus and 35S terminator sequences resulting in BASTA-resistant plants (Supplementary Data 1).The Cas9 and the two gNIA1 transcripts were either fused to TLS1 (tRNAMet sequence) or TLS2 (tRNAMet-ΔDT sequence, lacking the D and T loop)5 and a short poly-A tail was added to the 3′ end of each gNIA1-TLS (Fig. 1a,b and Supplementary Data 1). Co-fold RNA structure predictions11 confirmed that folding of the TLS motif was unlikely to be affected in the gRNA or Cas9 fusion constructs (Fig. 1b,c). Also, addition of related TLS motifs to gRNAs was shown not to interfere with the function of gRNAs12,13.
Cas9/gRNA-TLS fusions move from root to shoot in grafted Arabidopsis
We first tested whether Cas9 and/or gNIA1 transcripts were root-to-shoot mobile without a TLS by hypocotyl grafting Cas9 × gNIA1 rootstocks with wild-type (Col-0) scions. Three weeks after grafting, shoot and root samples were harvested separately (n = 4, samples were pools of 4–6 independent grafted plants) (Fig. 2a). We did not detect Cas9 or gRNA in these wild-type scion samples, though both were detected in the rootstock samples, indicating that Cas9 mRNA and gRNA transcripts targeting NIA1 are not root-to-shoot mobile.
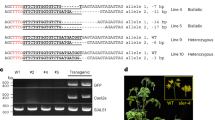
a, Three weeks after grafting shoot (S) and root (R) samples were harvested from plants cultured on 5 μM estradiol and 0% sucrose 0.5 MS medium. b, RT–PCR detection (45 PCR cycles) of Cas9 and gNIA1 transcripts (each sample a pool of 4–6 grafted plants) in transgenic root and grafted wild-type scion samples. Stars indicate presence of Cas9-TLS and gNIA1-TLS amplicons in Col-0 wild-type. M indicates molecular weight markers. RT–PCR of kanamycin (Kan) and hygromycin (Hyg) transcripts were used as contamination controls. Note that transcript absence was confirmed using 50 PCR cycles. c, Chlorotic leaf phenotype of nia1 chimeric mutant in juvenile grafted plants. Dashed boxes indicate chlorotic phenotype detected in wild-type leaves. TG1, Cas9 × gNIA1 (control); TG2, Cas9-TLS1 × gNIA1-TLS1; and TG3, Cas9-TLS2 × gNIA1-TLS2. d, Genomic PCR assays to detect edited NIA1 fragment with a deletion. PCR amplicons marked by stars indicate CRISPR–Cas9-induced mutations in wild-type tissue. Four independent replicates (4–6 pooled root or shoot samples) per graft combination were analyzed. Expected wild-type NIA1 amplicon is 1,469 bp and edited NIA1 is approximately 430 bp. e, Edits induced by mobile CRISPR–Cas9 confirmed by Sanger sequencing. Red bases indicate gNIA1 target 1 and gNIA1 target 2 sequences. Bold and underlined bases indicate the two protospacer-adjacent motif sites of the gRNA.
Conversely, in a similar experiment with rootstocks expressing Cas9-TLS1 and gNIA1-TLS1 or Cas9-TLS2 and gNIA1-TLS2, we detected the Cas9-TLS (four replicates for each TLS1 and TLS2 fusion) and the gNIA1-TLS (four replicates for each TLS1 and TLS2 fusion) transcripts in grafted transgene-free wild-type shoots (Fig. 2b). This finding confirmed that adding a TLS motif is sufficient to promote long-distance mobility of the Cas9 and gRNA transcripts. As a control for tissue contamination and specificity of TLS-mediated transport, we used the hygromycin and kanamycin resistance genes expressed by transgenic roots. Both resistance genes were not detected in the wild-type shoot samples indicating that no contamination occurred during harvest and that both transcripts are not root-to-shoot transmissible (Fig. 2b).
Even though the Cas9-TLS and the gNIA1-TLS transcripts were moving from root to shoot, low transcript abundance or lack of Cas9-TLS translation in distal tissues might result in insufficient or no editing activity. Therefore, we asked whether gene deletion edits are detectable in root and shoot tissue samples of grafted plants. A chlorotic nia1 mutant phenotype was detected in some leaves formed on grafted scions of plants grown on NH4-deficient medium 14 days after grafting (Fig. 2c), indicating that the mobile transcripts were functional in recipient wild-type tissues. This was observed in 20 out of 28 of Col-0/Cas9-TLS1 × gNIA1-TLS1 grafted plants and in 26 out 30 of Col-0/Cas9-TLS2 × gNIA1-TLS2 grafted plants, but never observed in the 20 tested Col-0/Cas9 × gNIA1 grafted plants. In line, NIA1 deletion edits could be detected in all grafted transgenic roots (Fig. 2d) and in all tested wild-type scions grafted onto the mobile transcripts producing Cas9-TLS1 × gNIA1-TLS1 (n = 4) and Cas9-TLS2 × gNIA1-TLS2 (n = 4) rootstocks (Fig. 2d,e). Serving as contamination controls, we also probed for the presence of kanamycin resistance gene, which was not detectable in genomic PCR assays performed on wild-type scion samples (Fig. 2b,d). These results also show that the root-produced mobile Cas9-TLS transcript was translated into a functional protein in recipient tissues. Further, this indicates that both Cas9-TLS and gNIA1-TLS are transported in sufficient amounts for effective genome editing of the NIA1 gene in grafted non-transgenic wild-type scions in the seedling stage.
Mobile Cas9/gRNA produces heritable gene edits
Previous studies suggested that a dominant-negative acting DISRUPTION OF MEIOTIC CONTROL 1 transcript interfering with meiosis fused to TLS motifs is graft transmissible from rootstocks to wild-type germline cells interfering with male sporogenesis in Nicotiana sp.5. Additionally, grafting experiments with mobile short interfering RNAs (siRNAs) showed that these are transported to meiotic precursor cells14. Therefore, we considered it to be likely that Cas9-TLS and gNIA1-TLS fusions can be delivered to wild-type flowers and cause heritable edits in the germline cells. Alternatively, these mobile fusions could be transported to meristems that would then give rise to a genome-edited lineage resulting in edited germline progenitor cells producing the next generation. To address this notion, we asked whether the Cas9-TLS and gNIA1-TLS fusion transcripts moved to leaves (cauline/rosette) and along the stem to reproductive tissues (siliques/flowers) in adult grafted plants. To detect the presence of the Cas9 and gRNA transcripts we performed PCR with reverse transcription (RT–PCR) assays on RNA samples from adult grafted plants (Fig. 3a,b). These assays indicate that Cas9 and gNIA1 lacking TLS motifs were not moving to distant tissues (n = 4; independent replicates for each tissue). By contrast, both Cas9 and gNIA1 fused to either TLS1 or TLS2 were detected in three out of four samples of grafted plants in scion tissues, respectively (Fig. 3b and Extended Data Fig. 1). These findings were confirmed by quantitative PCR with reverse transcription (RT–qPCR) assays. Here, with wild type/Cas9 × gNIA1 grafts, we do not detect Cas9 transcripts above technical background noise levels in wild-type siliques, flowers, stem, cauline, and rosette leaves (Fig. 3c). By contrast, Cas9-TLS1 × gNIA1-TLS1/wild type and Cas9-TLS2 × gNIA1-TLS2/wild type grafted plants, we detect Cas9-TLS transcripts in wild-type siliques, flowers, stem, cauline leaves, and rosette leaves. We estimated the transcript root-to-shoot delivery ratio using the RT–qPCR data, which indicates that approximately 1 out of 1,000 root-produced transcripts is delivered to non-transgenic shoot tissues (Table 1 and Fig. 3c). Here no significant difference was detected between Cas9-TLS1 and Cas9-TLS2 presence in the scion samples. We next analyzed whether the delivered fusion constructs are active by confirming the presence of genome edits in wild-type siliques and flowers by genomic PCR and Sanger sequencing of independent replicates (Fig. 3d,e and Extended Data Fig. 2). These assays revealed the presence of the expected NIA1 genomic deletions in recipient wild-type tissues. This indicates that the delivered Cas9-TLS transcripts were translated into a functional protein in recipient tissues, and that both Cas9-TLS and gNIA1-TLS transcripts are delivered in sufficient amounts for effective genome editing of the NIA1 gene in non-transgenic tissue of adult plants.
a, Appearance of grafted plants (43 days after grafting) grown on soil and treated with 5 μM estradiol. b, RT–PCR detection (45 PCR cycles) of Cas9 and gNIA1 transcripts in rootstock (R) and grafted wild-type tissues samples from silique (Sil), flower (Flo), cauline leaf (Cau), stem (St), and rosette (Ro). Four independent replicates (each sample a pool of three grafted plants) were analyzed. Stars indicate RT–PCR Cas9-TLS and gNIA1-TLS amplicons detected in wild-type scion samples. Kan and Hyg transcripts serve as contamination controls. Note that transcript absence was confirmed by 50 PCR cycles. c, RT–qPCR detection of mobile Cas9-TLS transcripts in scion tissues and in grafted rootstock (Root). y-axis, mean \(2^{-\Delta\Delta c_t}\) values; log10 scale. Each value represents the mean of three independent biological replicates presented as black dots on the bar. Note that every \(2^{-\Delta\Delta c_t}\) data point below the dashed line represents technical background as measured with wild-type scion samples grafted onto wild-type rootstocks with no Cas9 transcript present. Significance was calculated using Student’s t-test (two tails); P values indicated by a, b, and c: a,b = 8.31567 × 10−22; b,c = 1.16396 × 10−22; a,c = 1.90267 × 10−11. d, Genomic PCR to detect NIA1 edited fragments in siliques and flowers. Samples were from the same plant material as analyzed in c. e, Mobile CRISPR–Cas9-induced genome edits confirmed by Sanger sequencing.
For the grafted plants, we next allowed the wild-type scions to set seeds, which were germinated on NH4-deficient medium to evaluate the appearance of nia1 homozygous chlorotic leaf and small size phenotypes (Fig. 4a), and the presence of NIA1 gene edits by genomic PCR and Sanger sequencing (Fig. 4b,c). As a nia1 mutant phenotype is most noticeable when a plant is homozygous, we concluded that we were underestimating the frequency of genome editing inherited by the progeny. Therefore, we also examined the seedlings for the presence of the edited NIA1 gene by genomic PCR to calculate the frequency of heritable genome edits created by graft-mobile Cas9 and gNIA1 (Fig. 4d, Extended Data Fig. 3 and Supplementary Table 1). In total 11 out of 15 grafts with Col-0 (wild-type) scions and Cas9-TLS1 × gNIA1-TLS1 rootstock and 17 out of 22 grafts with Col-0 scions and Cas9-TLS2 × gNIA1-TLS2 rootstock produced offspring with detectable genome edits. No NIA1 edits were found in the progeny of 11 wild-type scions grafted on Cas9 × gRNA lines lacking the TLS sequences. The calculated frequency of editing induced by mobile Cas9-TLS and gNIA1-TLS transcripts was between 5.7 (TLS1) and 5.0 (TLS2) per 1,000 seeds of which 1.17 (TLS1) and 1.41 (TLS2) per 1,000 seeds showed a clear nia1 mutant phenotype indicating that these were homozygous (Fig. 4d). The TLS1 or TLS2 constructs caused no significant editing frequency difference according to Student’s t-test (two tails, P > 0.1) suggesting that both are similarly active in delivering editing factors to recipient tissues.
a, Screening for NIA1 gene-edited grafted plants offspring and phenotyping for nia1 homozygous mutants generated by mobile CRISPR–Cas9. Homozygotic nia1 mutants were phenotypically screened from 14-day-old seedlings grown on NH4-limited plates and submitted to PCR assays. Scale bar, 5 mm. b, Genomic PCR to detect NIA1 edits in the wild-type scion progeny. Edited NIA1 PCR amplicons are marked by stars. All samples were harvested from 10-day-old seedlings grown on NH4-limited plates. Each replicate is a pool of ~70 to ~100 seedlings (Supplementary Table 1). Cas9 and Kan transcript RT–qPCRs were used to confirm that the progeny was transgene-free. c, Transgene-free genome edits confirmed by Sanger sequencing. d, Editing efficiency analysis of Cas9-TLS/gRNA-TLS constructs in the progeny of grafted plants (Supplementary Table 1). Asterisk indicates the number of homozygotes identified in 1,000 seedlings.
To further confirm whether this graft-mobile gene editing system also works on other gene targets and thus in the context of other gRNA targeting sequences, we created two gRNAs, gVenus1 and gVenus2, with and without fused TLS1 or TLS2 sequences. These gRNAs target an artificial 35Spromoter::H2B-Venus::35Sterminator::BastaR construct (Extended Data Fig. 4a and Supplementary Data 1). After accurate double editing by both gVenus1 and two gRNAs, this artificial construct should lose the H2B-Venus::35Sterminator sequences and consequently express the downstream BastaR resistance gene. Offspring of such gene-edited plants should gain dominant BASTA resistance and homozygous plants should lose Venus yellow fluorescence.
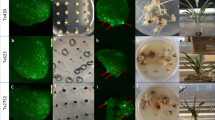
We grafted rootstocks expressing Cas9 and gVenus with and without TLS fusions with 35Spromoter::H2B-Venus::35Sterminator::BastaR transgenic scions and examined the offspring (seedlings) for the presence of the edited Venus gene by genomic PCR and in part by Sanger sequencing to calculate the frequency of heritable genome edits created by graft-mobile Cas9 and gVenus (Extended Data Figs 4b and 5, and Supplementary Table 2). In total 19 out of 20 grafts with 35Spromoter::H2B-Venus::35Sterminator::BastaR transgenic scions and Cas9-TLS1 × gVenus-TLS1 rootstock and 17 out of 18 grafts with 35Spromoter::H2B-Venus::35Sterminator::BastaR transgenic scions and Cas9-TLS2 × gVenus-TLS2 rootstock produced offspring with detectable genome edits. Again, no edits were found in the progeny of 19 wild-type scions grafted on Cas9 × gVenus lines lacking the TLS sequences. The calculated frequency of editing induced by mobile Cas9-TLS2 and gVenus-TLS2 transcripts was above 15.9 per 1,000 seeds (Extended Data Fig. 4d and Supplementary Table 2). However, none of the edited offspring showed the anticipated BASTA resistance. This was because the gVenus1 gRNA cut the 35Spromoter target region further upstream as predicted which resulted in a truncated 35S promoter (Extended Data Fig. 4a,b). Thus, the deletion edit did not activate expression of the downstream BastaR gene providing resistance. We next analyzed the presence of Venus fluorescence in the offspring of three homozygotic 35Spromoter::H2B-Venus::35Sterminator::BastaR scions grafted on Cas9-TLS2 × gVenus-TLS2 rootstocks. Here we detected loss of Venus fluorescence in 7 of 1,557 offspring seedlings indicating presence of homozygous gene edits in approximately 0.45% of plants (Extended Data Fig. 4c,d). In summary, these results indicate that the mobile CRISPR–Cas9 transgene-free genome editing is also functional with alternative gRNA sequences and that without the addition of TLS motifs it is not introducing heritable gene edits in grafted wild-type scions.
Cas9/gRNA-TLS moves from Arabidopsis roots to Brassica rapa shoots
We next asked whether the mobile Cas9-TLS and gRNA-TLS constructs are also transported to a distantly related crop species grafted onto transgenic Arabidopsis roots. To address this, we grafted Brassica rapa, a worldwide cultivated vegetable and oilseed crop, with the Arabidopsis lines expressing the mobile and non-mobile NIA1-targeting gRNAs. As the NIA1 gene sequence is conserved between Arabidopsis and Brassica rapa we would expect to detect Cas9-TLS and gRNA-TLS transcripts and NIA1 edits in shoots of grafted B. rapa/Cas9-TLS × gNIA-TLS plants. To test this, we hypocotyl-grafted Arabidopsis Cas9 × gNIA1 or Cas9-TLS2 × gNIA1-TLS2 rootstocks with B. rapa scions (Fig. 5a) and confirmed successful healing of the graft junction 10, 20, and 40 days after grafting by slightly pulling the scion and rootstock. This cross-species hypocotyl grafting protocol was very successful with 13 out of 15 B. rapa/Cas9 × gNIA1 and 15 out of 16 B. rapa/Cas9-TLS2 × gNIA1-TLS2 grafted plants passing the graft take test.
a, Appearance of grafted plants (20 and 40 days after grafting) grown on MS medium and treated with 5 μM estradiol. Red arrows indicate the graft junction. b, RT–PCR detection (45 PCR cycles) of Cas9 and gNIA1 transcripts in rootstock, and grafted wild-type tissues samples from silique, flower, leaf, and stem from one independent replicate (the other three replicates are presented in Extended Data Fig. 3) was analyzed. Stars, RT–PCR Cas9-TLS and gNIA1-TLS amplicons detected in B. rapa wild-type scion samples. Kan and Hyg transcripts serve as contamination controls. Note that transcript absence was confirmed by 50 PCR cycles. c, RT–qPCR detection of mobile Cas9-TLS transcripts in scion tissues and in grafted rootstock (Root). y-axis, mean \(2^{-\Delta\Delta c_t}\) values; log10 scale. Each value represents the mean of three independent biological replicates presented as black dots on the bar. Note that every \(2^{-\Delta\Delta c_t}\) value below the dashed line represents technical background as measured with wild-type scion samples grafted onto wild-type rootstocks with no Cas9 transcript present. Significance was calculated using Student’s t-test (two tails); P values indicated by a, b and c: a,b = 3.48073 × 10−8; b,c = 2.29801 × 10−7; a,c = 3.43932 × 10−8. d, Genomic PCR to detect NIA1 edited fragments in siliques and flowers. Samples were from the same plant material as analyzed in b,c. e, Mobile CRISPR–Cas9-induced genome edits confirmed by Sanger sequencing.
We next asked whether Cas9 and/or gNIA1 transcripts, with or without a TLS transport motif are also root-to-shoot mobile in these heterografted plants. We sampled RNA and gDNA from siliques, flowers, stem, leaves, and roots (n = 4, two grafted plants pooled for each sample) 40 days after grafting and submitted the samples to RT–PCR and genomic PCR assays (Fig. 5a). In B. rapa scion samples from plants grafted on Arabidopsis Cas9 × gNIA1 transgenic roots we did not detect Cas9 or gRNA transcripts (n = 4, each sample pool of two grafted plants), although both were present in the rootstock samples (Fig. 5b,c and Extended Data Fig. 6). By contrast, in the B. rapa siliques, flowers, stem, and leaf samples harvested from plants grafted onto Cas9-TLS2 × gNIA1-TLS2 rootstocks, we detected both Cas9-TLS2 and gNIA1-TLS2 (n = 4, each sample pool of two grafted plants) (Fig. 5b,c and Extended Data Fig. 6). We next measured the Cas9 transcript levels in B. rapa siliques, flowers, stem, leaf, and Arabidopsis root tissue samples by RT–qPCR. In B. rapa samples harvested from plants supported by Cas9 × gNIA1 rootstocks no Cas9-specific transcript could be detected. In B. rapa samples from plants supported by Cas9-TLS2 × gNIA1-TLS2 rootstocks we found relatively high levels of Cas9-TLS2 transcripts (Fig. 5c) as indicated by the higher shoot/root ratio of ~0.4 (Table 1) compared to homografted Arabidopsis plants showing a ratio of ~0.09 (Table 1). Thus, the TLS motif was efficiently promoting long-distance mobility of the Cas9 and gRNA transcripts from Arabidopsis rootstocks to B. rapa scions. We also tested whether mobile Cas9-TLS and gRNA-TLS fusions are functional in introducing deletion edits into the conserved B. rapa NIA1 gene. Consistent with the findings with heterografted Arabidopsis, we detected genome edits in four out of six siliques and in four out of six flowers formed on grafted B. rapa plants by genomic PCR and Sanger sequencing (Fig. 5d,e and Extended Data Fig. 7). These findings suggest that both Cas9-TLS and gNIA1-TLS constructs are transported in sufficient amounts and functional in editing the genome of a heterografted crop plant.
Discussion
We have demonstrated a simple, effective method for creating heritable transgene-free genome edits using grafted plants. TLS motifs fused to Cas9 and gRNA mediate their transport from transgenic roots to grafted wild-type shoots. The grafted recipient wild-type tissues show edited genomes and their flowers produce seeds with inherited edited genomes. The detected efficiency of inherited deletion edits in the range of ~0.1% for homozygotic and of ~1.6% for heterozygotic edits was reasonably high, although these numbers are most likely an underestimation as pools of approximately 100 or 40 seedlings for NIA1 or Venus edits were analyzed, respectively (Fig. 4d and Extended Data Fig. 5). Indeed, as a deletion depends on two gRNA edit sites, the actual editing efficiency, including single position edits, induced by mobile Cas9 and gRNAs in wild-type scions, is most likely underestimated. Given that multiplex PCRs allow for fast screening of edits in a large number of seedlings deriving from grafted plants, homozygous mutants can be identified in a relatively short time. Notably, the presented approach is not only useful for plant researchers to generate stable transgene-free lines harboring, for example, multiplex genome edits15, but also for more complex applications. For example, as not all somatic cells will be edited in grafted wild-type scions, this also constitutes a chimeric somatic mutant allowing for the analysis of otherwise lethal mutations.
The zCas9 construct used in this study, with approximately 4,200 bases in length, is currently the longest mRNA that has been made mobile by the addition of an RNA mobility motif, and Cas9-TLS is one of the largest mobile mRNAs reported for Arabidopsis16. Further, our data show that mobile Cas9-TLS and gRNA-TLS transcripts are delivered to heterologous plants and that root-produced Cas9-TLS transcripts are indeed translated into a functional editing enzyme in recipient scion cells. We also noticed that approximately 1 of 250 Arabidopsis-root-produced Cas9-TLS transcripts is detectable in B. rapa scion tissues as estimated from RT–qPCR data (Table 1 and Extended Data Fig. 5c), implying that a reasonably high number of Cas9-TLS transcripts are actually delivered over graft junctions. While the exact mechanisms of TLS-induced mRNA mobility from roots to shoots remain to be elucidated, we have demonstrated that mobile TLS fusion constructs move to all major shoot tissues such as rosette and cauline leaves, stem, flowers, and seed-bearing siliques (Fig. 3b) where they can induce genome editing. We also noticed that, although siliques seem to receive lower amounts of mobile Cas9-TLS transcript, they appear to show more genomic deletions than flowers or rosette leaves (Extended Data Fig. 2). We hypothesize that the appearance of more gene edits in the seed producing siliques is either due to their clonal origin from flower meristems or the capacity of meiocytes to receive graft-mobile transcripts, as seen with gene-silencing-inducing siRNAs14. In line, it is known that addition of a TLS motif can promote long-distance mRNA mobility in Arabidopsis and Nicotiana tabacum. In both species, a dominant-negative acting DISRUPTION OF MEIOTIC CONTROL 1 (DNDMC1) variant, which lacks the N-terminal 92 amino acid residues and therefore interferes with meiotic progression, fused to TLS is transported from grafted rootstocks to wild-type flowers and interferes effectively with meiosis in recipient meiocytic cells5. Other mobile RNA motifs, derived from FLOWERING LOCUS T (FT) transcripts, when fused to gRNAs seem to license transport of viral expressed gRNAs in A. thaliana, Nicotiana sp., and wheat12,17. Therefore, RNA mobility mediated by RNA transport motifs seems to be a conserved mechanism which can be used across a wide range of plant families and crop species. Although it seems unlikely and has to be experimentally shown, one could speculate that Cas9 mobility might be increased by adding multiple TLS motifs or alternative relatively short RNA mobility motifs identified in other mobile mRNAs, such as TRANSLATIONALLY CONTROLLED TUMOR PROTEIN 1 (TCTP1), that mediate transport of heterologous transcripts18.
The major task in gene editing of plants is to generate transgene-free edited offspring to ensure genomic stability and their use in food production. For this purpose, one must eliminate all editing components from the edited crop plant before their commercial release. Thus, elimination of exogenous DNA components has become one of the major objectives in gene editing research. This can be achieved by local expression of Cas9/gRNA-related gene components using Agrobacterium-mediated transformation19,20 or plant-virus-mediated gene editing21,22, or by directly delivering active Cas9 protein–gRNA complexes into plant cells18,23,24,25,26. The disadvantages of these systems are that many crop plants and fruit trees are inaccessible or that they are difficult to be transiently infected or transformed. For example, in the previous methods the edited plant material still contains the Cas9 gene and/or remains contaminated by transgenic bacteria or viruses. This is undesirable for commercial crop production owing to expensive containment and time-consuming elimination protocols that have to be implemented and the risks involved by using genetically modified viruses in production pipelines.
Here we present an alternative, rapid method for producing heritable genome edits induced by graft-mobile TLS fusion transcripts. The presented grafting system does not need elimination of gRNA or Cas9 transgenes and regeneration of plants from tissues or protoplasts and, thus, the desired mutation can be produced and used after the first generation. Alternative approaches resulting in transgene-free edited plant material rely either on delivery of active Cas9–gRNA protein–RNA complexes into callus cells, protoplasts, or immature embryos followed by elaborative and time-consuming regeneration and selection of edited plant material for further use. Again, this is difficult or not possible for most crop species. In the presented grafting system, Cas9-TLS mRNA and gRNA-TLS fusions are delivered directly to germline progenitor cells where they are actively editing the genome. This allows for producing edited seeds with no need for transgene or virus elimination.
The gene editing method presented here, despite the required production of a transgenic rootstock line, is not more time consuming than alternative transgene-free genome editing methods. Gene editing by grafting confers an advantage as there is no requirement for multiple additional generations to remove the transgene or regeneration of plants from transfected protoplasts. For example, by our estimates, it takes approximately 3 months to generate a T1 transgenic Arabidopsis rootstock line that can be used as a gene editing donor, another 3 to 4 months to generate seeds and to select by genomic multiplex PCR individual edited lines. In comparison to an alternate transgene-free editing method using protoplast transfection with pre-assembled ribonucleoprotein complexes26, we find that the time required is similar—approximately 3 months to generate edited plantlets, then an additional 3 to 4 months to produce and select the next generation of seeds. While that method is efficient, one should also consider that protoplast regeneration protocols do not exist for a wide variety of plant species and that protoplast regeneration has been observed to induce genome instability27, which has to be assessed in the following generations. Though both the grafting method and the protoplast method each have advantages and disadvantages, the total time required for both methods is similar. The grafting system also permits for the introduction of edits in any graft-compatible cultivar that can be combined with a given Cas9-TLS/gRNA-TLS expressing rootstock. While the production of a transgenic rootstock is time consuming, any rootstock species that is graft-compatible and easily transformable can be used as a donor for the graft-mobile gene editing system. Also, once established, a rootstock line can be easily maintained and distributed and used as a donor plant line for any variety or graft-compatible crop species of particular interest for breeders. That this is feasible we have shown by using transgenic Arabidopsis rootstocks as donors to introduce gene edits to grafted B. rapa scions that show different growth and flowering behavior.
As mentioned above, the introduction of gene edits by grafting would greatly simplify the production of mutant plants where transformation is difficult, impossible or undesirable. The wild-type scion cultivar grafted onto a given transgenic rootstock donor can belong to a very distant plant family. For example, A. thaliana and Nicotiana sp. can easily be used as rootstocks for a very wide range of quite distantly related species including tomato, carrots, soybean, and onions28. Although both species are good donor plants that can be grafted with many crop species, there is still a limitation for their use with distantly related species. One has to consider that the donor (stock) produced gRNA sequences must be optimized to match the gene sequences of more distantly related species to warrant sufficient sequence similarity for successful editing. Thus, it might be required to produce additional transgenic donor lines expressing optimized gRNAs sequences specifically matching the target gene(s) of heterologous scions.
In light of the recent study that demonstrates grafting in monocotyledonous species29, we can further speculate that this technique based on mobile Cas9-TLS and gRNA-TLS fusions will find use in main crops such as maize, wheat, and rice. In addition, a graft-transmissible editing system could be used in many agricultural settings as grafting is a major technological platform in commercial agricultural productions30.
Methods
Plasmid construction
The rbcs-e9t terminator sequence was PCR amplified from pJF3101 (a modified version of pHEE2E-TRI)10, using primers rbcsT FP and rbcsT RP (Supplementary Table 3). The PCR product was digested with BsaI to produce two XhoI-compatible ends, such that the 3′ end creates a non-functional scar site in place of the XhoI site, leaving the final vector with one remaining XhoI site at the 5′ end of the rbcs-e9t sequence. This was cloned into the estradiol-inducible binary vector pMDC731 at the XhoI site, and the orientation of insert was confirmed by digestion with XhoI and AscI. This intermediate vector was termed pMDC7_rbcs-e9t.
We used the Cas9 (zCas9) sequence from pJF103110. This Cas9 sequence features an optimized codon usage for plants, a nuclear localization signal, and a FLAG-tag at the 3′ end of the Cas9 coding sequence. The Cas9 and Cas9-TLS fusion sequence fragments were made by PCR amplification using Cas9 FP and Cas9 RP, Cas9-ΔDT RP or Cas9-tMet RP primers (Supplementary Table 3) containing BsaI sites with compatible overhangs (Extended Data Fig. 8). Amplification was done using Phusion DNA polymerase (Thermo Fisher) with the addition of 3% dimethylsulfoxide to prevent folding of the TLS primers. Amplified regions were cloned into CloneJet Blunt end ligation vector (Thermo Fisher), confirmed by sequencing, and cloned into the XhoI site in pMDC7_rbcs-e9t binary gateway vector using the compatible BsaI sites at the 5′ and 3′ end. The pMDC7 Cas9 clones were transformed into DB3.1 E. coli strain owing to the toxicity of the ccdb gene present in pMDC730,31.
The gRNA backbone sequences Fragment 1 and Fragment 2 were created by gene synthesis (Eurofins) (Extended Data Fig. 8). Fragment 1 (with or without TLS motifs) was cloned into pENTR4 using BamHI and XhoI. Fragment 2 sequences were used as PCR templates for amplification with NIA1 and Venus target sequence primers with BsaI cut sites (NIA1 Fragment 2 FP/RP and Venus Fragment 2 FP/RP; Supplementary Table 3). Fragment 2 (with NIA1 and Venus target sequences added) were cloned into Fragment 1 using BsaI restriction sites with compatible overhangs. Completed gRNA (gNIA1 and Venus target 1 and 2) expressing sequences in pENTR4 were then transferred into pMDC10031 by Gateway cloning (LR reaction). The 35Spromoter::H2B-Venus:35Sterminator:Basta:Nosterminator binary plasmid was constructed by gene synthesis and entry vector gateway cloning into the pRI 909 destination vector. All final constructs in the binary vectors were confirmed by Sanger sequencing and are listed in Supplementary Table 4.
Generation of transgenic arabidopsis lines
Binary vector constructs were transformed to Agrobacterium tumefaciens strain AGL1 subsequently used to transform Arabidopsis Col-0 by the floral dip method32. T1 seedlings were selected on 0.5 MS, 0.5% sucrose plates solidified with 0.68% micro agar (Duchefa Biochemie). gNIA1, gNIA1-TLS1, gNIA1-TLS2, gVenus-TLS1, or gVenus-TLS2 expressing transgenic lines were crossed with transgenic lines harboring estradiol-inducible Cas9, Cas9-TLS1, or Cas9-TLS2 sequences, respectively. The antibiotics used for each construct can be found in Supplementary Table 2. Lines were screened for 3:1 segregation on antibiotic selection medium and the absence of visible growth phenotypes.
Hypocotyl grafting of arabidopsis with arabidopsis
Arabidopsis thaliana Col-0 wild-type and transgenic seeds were placed on plates containing 0.5 MS salts, 1% sucrose, and 1% micro agar (Duchefa Biochemie) and vertically grown in short-day conditions (8 h light/16 h dark; day 22 °C/night 19 °C; light intensity: 170 μE m−2 s−1). Seedlings (6–7 days after germination, DAG) with ~4 cm long roots were cut in the upper half of the hypocotyl using a sterile razor blade. Silicon micro-tubes (0.3 mm internal diameter) were used to stabilize the graft junction. Grafted plants were transferred onto new plates (0.5 MS salts, 0% sucrose, 1% micro agar (Duchefa Biochemie), and 5 µM estradiol) and vertically grown in short-day conditions. Beginning at 6–7 days after grafting, adventitious roots appearing on the upper (scion) hypocotyl junction were removed every day. Twenty-one days after grafting (young) plants were harvested from plates for RNA and DNA isolation, or 14 days after grafting plants were transferred to soil with 5 µM estradiol (16 h light/8 h dark), 43 days after grafting (adult flowering) plants were harvested for RNA and DNA isolation, or kept to produced seeds.
Hypocotyl grafting of B. rapa with Arabidopsis
Sterilized Arabidopsis thaliana Col-0 transgenic seeds were placed on plates containing 0.5 MS salts, 1% sucrose, and 1% micro agar (Duchefa Biochemie) and vertically grown in short-day conditions (8 h light / 16 h dark; day 22 °C / night 19 °C; light intensity: 170 μE·m − 2·s−1). 7 days after preparing Arabidopsis seeds, sterilized B. rapa (RCBr) wild-type seeds (provided by the John Innes Centre, UK) were placed on plates containing 0.5 MS salts, 1% sucrose, and 1% micro agar (Duchefa Biochemie) and vertically grown in long-day conditions (16 h light/8 h dark; day 22 °C/night 19 °C; light intensity: 170 μE m−2 s−1). Two-week-old Arabidopsis seedlings and one-week-old B. rapa seedlings were used for grafting. Arabidopsis plants were cut in the lower half of the hypocotyl for use as a rootstock. To prepare B. rapa scions for grafting the cotyledons were removed and then the hypocotyl was cut 2 cm below the shoot apical region. The Arabidopsis rootstock and B. rapa scion were aligned together on a new plate containing 0.5 MS salts, 1% sucrose, and 1% micro agar (Duchefa Biochemie). Grafted plants were grown under long-day conditions with plates in a vertical orientation (5–10°). The graft success was evaluated 10 and 14 days after grafting by gently pulling the scion and rootstock apart. Fourteen days after grafting plants were transferred to new 0.5 MS plates (1% sucrose, and 1% micro agar) supplemented with 5 µM estradiol to induce gene expression in the root and grown under long-day conditions (16 h light/8 h dark; day 22 °C/night 19 °C; light intensity: 170 μE m−2 s−1). Twenty-five days after grafting plants were transferred to 0.5 MS agar medium (1% sucrose, supplemented with 5 µM estradiol) in glass jars and grown till flowering (approximately 40 days after grafting) and harvested for RNA and genomic DNA isolation (Fig. 5).
Genomic DNA extraction and genomic edits detection by PCR
Plant tissue was harvested, frozen in liquid nitrogen, and stored at −80 °C in Eppendorf tubes until extraction. Frozen tissue was incubated in DNA extraction buffer, manually broken, and centrifuged to remove debris. The supernatant was transferred to a new tube and extracted with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, v/v), homogenized and centrifuged for 10 min (13,000g). The upper phase was transferred to a new tube, mixed with four volumes of cold isopropanol, and DNA was precipitated at −20 °C overnight and collected (15 min, 13,000g). The DNA pellet was washed twice with ice-cold 70% ethanol and resuspended in 25 µl distilled water.
Specific PCR primers for NIA1 deletion edits (NIA1 Edit Detection FP/RP) and VENUS deletion edits (VENUS Edit Detection FP/RP; Supplementary Table 1) were used to amplify the region surrounding the predicted targeted edit site, producing a 1,469 bp (NIA1) and 1,719 bp (Venus) band in the wild-type genome with approximately 430 bp and 250 bp in the edited (deletion mutant) genome, respectively. Fifty nanograms of gDNA was used for a 20 μl PCR reaction with Phusion DNA Polymerase (NEB #M0530). PCR annealing temperature of 55 °C and extension time of 5 s was used to promote amplification of the smaller gene-edited fragment. Forty-five PCR cycles were used unless otherwise stated. To confirm the identity of bands, PCR fragments were isolated from the gels and ligated into CloneJet blunt-end ligation vector (Thermo Fisher) before being transformed to E. coli. Twenty clones were selected for each graft and were submitted to Sanger sequencing (LGC Genomics).
RNA isolation and RT–PCR
Plant tissues, either from juvenile grafted plants or from adult flowering grafted plants were independently collected in 2-ml Eppendorf tubes containing metal beads. After freezing in liquid nitrogen, 750 µl TRIzol Reagent (Invitrogen) was added before samples were homogenized and incubated at room temperature for 5 min. Chloroform was then added (0.2 ml per 0.75 ml TRIzol) and homogenized by vortexing. The upper (aqueous) phase was transferred to 1.5-ml tubes and 1 volume of isopropanol and 0.1 volumes of 3 M NaOAc (pH 5.2) were added to precipitate total RNA. RNA pellets were then washed twice with 80% ethanol and then once with 99% ethanol. The RNA pellets were resuspended in 25 µl DEPC-treated water.
Reverse transcription was done using AMV Reverse Transcriptase (Promega). Approximately 1.5 µg isolated total RNA was used per reaction. Oligo(dT) primer was used for cDNA production of mRNAs including Cas9, kanamycin, hygromycin, and ACTIN2, a gRNA-specific primer (gRNA RT-RP; Supplementary Table 2) was also included for reverse transcription of gNIA1 gRNAs to facilitate detection by PCR. The total RNA was denatured at 70 °C for 10 min followed by annealing incubation for 5 min at 37 °C. The RT reaction was done at 42 °C for 90 min followed by inactivation at 72 °C for 10 min.
RT–PCR was done using Phusion polymerase according to the standard protocols. ACTIN2 primers (Supplementary Table 2) and ACTIN2 amplification was done with 30 PCR cycles and to confirm cDNA quality. PCRs with primers for detecting Cas9 and gRNA mobility were done using 45 PCR cycles for increased sensitivity. To confirm signal (amplicon) absence 50 PCR cycles were used unless specified otherwise in the text. As a control for tissue contamination and specificity of transport, we used the hygromycin and kanamycin resistance genes as targets for RT–PCR as these transcripts were shown to be not root-to-shoot graft transmissible in this study.
RT–qPCR
To measure Cas9 transcript levels by RT–qPCR we used an Applied Biosystems 7900HT fast Real-time PCR machine with SYBR Green as readout following the supplier’s manual. For all assays three technical replicates were performed. The program of thermal cycling was as follows. Step 1: 1 cycle, 2 min at 50 °C. Step 2: 1 cycle, 10 min at 95 °C. Step 3: 40 cycles, 15 s at 95 °C, 1 min at 60 °C. Dissociation step: 15 s at 95 °C, 15 s at 60 °C, 15 s at 95 °C. UBQ10 gene was used as the reference. Cas9 transcript expression levels presented by \(2^{-\Delta\Delta c_t}\) (ΔΔct = ∆ct (Cas9) − ∆ct (control average)); ∆ct (Cas9) = ct (Cas9) − ct (UBQ10); ∆ct (control) = ∆ct (∆ct average of wild-type grafted scions) following established protocols33. The bar plots in Fig. 3c were created using the GraphPad Prism9 software. Cas9 and UBQ10 qPCR primer sequences used see Supplementary Table 3.
nia1 Phenotype screening
Progeny seeds from grafted plants were surface sterilized by washing in 70% ethanol followed by washing in 100% ethanol and allowed to dry. Seeds were placed on NH4-free medium (see Supplementary Table 4), stratified at 4 °C for 2 days, and transferred to a growth chamber for germination (16 h light/8 h dark; day 22 °C/night 22 °C). Seedlings were grown for 14 days, and candidate seedlings showing a nia1 phenotype were transferred to new plates containing 0.5 MS, 1% sucrose, and 1% micro agar (Duchefa Biochemie) (Supplementary Table 5) for 2 weeks before being transferred to soil. Leaf pieces were then harvested for genomic DNA extraction and genomic DNA was used for PCR assays to confirm the presence of gene edits.
Measurement of genome edit efficiency
F1 seeds were sterilized as described above and sown on 0.5 MS medium containing 1% sucrose and solidified with 0.68% micro agar. Plates were stratified at 4 °C for 2 days before transfer to a growth chamber (16 h light/8 h dark; day 22 °C/night 22 °C). Ten days after germination, seedlings were harvested for genomic DNA isolation. Approximately 70 to 100 (for NIA1 edits) or approximately 40 (for Venus edits) seedlings were pooled together (Supplementary Table 1) and genomic DNA extracted as described above, except the DNA pellets were resuspended in 100 µl distilled water. Samples were then diluted by a factor of 50 before PCR, owing to the high concentration of DNA in the samples. To facilitate detection of an NIA1 and Venus deletion edits, we digested 2 µl of pooled genomic DNA samples with FastDigest HindIII (NIA1) or PstI (Venus) (Thermo Fisher) enzymes targeting the respective wild-type (non-edited) genomic DNA for 1 h at 37 °C in a 10 µl reaction using 1 µl (10 U) of enzyme following the manufacturer’s protocol. 2 µL of the digested sample was used for PCR using NIA Edit Detection FP / RP or Venus Edit Detection FP / RP primers as described above. PCR results from samples were then scored for the presence or absence of the edited NIA1 or Venus gene (Extended Data Figs 3 and 5), and partially confirmed by Sanger sequencing (Fig. 4c and Extended Data Fig. 6b). This information was then used to calculate the approximate minimal number of edited seedlings in the F1 generation as shown in Fig. 4d, Supplementary Tables 1 and 2, Extended Data Fig. 6c.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information). Source data are provided with this paper.
References
Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR–Cas9. Science 346, 1258096 (2014).
Mali, P., Esvelt, K. M. & Church, G. M. Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963 (2013).
Manghwar, H., Lindsey, K., Zhang, X. & Jin, S. CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125 (2019).
Kumagai, Y. et al. Introduction of a second ‘Green Revolution’ mutation into wheat via in planta CRISPR/Cas9 delivery. Plant Physiol. 188, 1838–1842 (2021).
Zhang, W. et al. tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28, 1237–1249 (2016).
Park, S. Y., Shimizu, K., Brown, J., Aoki, K. & Westwood, J. H. Mobile host mRNAs are translated to protein in the associated parasitic plant cuscuta campestris. Plants (Basel) 11, 93 (2021).
Cheng, C. L., Dewdney, J., Nam, H. G., den Boer, B. G. & Goodman, H. M. A new locus (NIA 1) in Arabidopsis thaliana encoding nitrate reductase. EMBO J. 7, 3309–3314 (1988).
Wang, R. et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136, 2512–2522 (2004).
Shan, Q. et al. Targeted genome modification of crop plants using a CRISPR–Cas system. Nat. Biotechnol. 31, 686–688 (2013).
Wang, Z. P. et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015).
Proctor, J. R. & Meyer, I. M. COFOLD: an RNA secondary structure prediction method that takes co-transcriptional folding into account. Nucleic Acids Res. 41, e102 (2013).
Ellison, E. E. et al. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 6, 620–624 (2020).
Xie, K., Minkenberg, B. & Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl Acad. Sci. USA 112, 3570–3575 (2015).
Zhang, W. et al. Graft-transmissible movement of inverted-repeat-induced siRNA signals into flowers. Plant J. 80, 106–121 (2014).
Xing, H. L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014).
Thieme, C. J. et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1, 15025 (2015).
Li, T. et al. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 14, 1787–1798 (2021).
Liu, W. et al. Lipofection-mediated genome editing using DNA-free delivery of the Cas9/gRNA ribonucleoprotein into plant cells. Plant Cell Rep. 39, 245–257 (2020).
Chen, L. et al. A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic. Res 5, 13 (2018).
Danilo, B. et al. Efficient and transgene-free gene targeting using Agrobacterium-mediated delivery of the CRISPR/Cas9 system in tomato. Plant Cell Rep. 38, 459–462 (2019).
Ali, Z., Eid, A., Ali, S. & Mahfouz, M. M. Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res 244, 333–337 (2018).
Hu, J. et al. A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol. Plant Pathol. 20, 1463–1474 (2019).
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K. & Mark Cigan, A. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274 (2016).
Zhang, Y. et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617 (2016).
Toda, E. et al. An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 5, 363–368 (2019).
Woo, J. W. et al. DNA-free genome editing in plants with preassembled CRISPR–Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164 (2015).
Fossi, M., Amundson, K., Kuppu, S., Britt, A. & Comai, L. Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 180, 78–86 (2019).
Notaguchi, M. et al. Cell-cell adhesion in plant grafting is facilitated by beta-1,4-glucanases. Science 369, 698–702 (2020).
Albacete, A. et al. Unravelling rootstock×scion interactions to improve food security. J. Exp. Bot. 66, 2211–2226 (2015).
Bernard, P., Gabant, P., Bahassi, E. M. & Couturier, M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene 148, 71–74 (1994).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 (2001).
Acknowledgements
We are grateful for the technical support of D. Schindelasch and C. Hauptvogel in our research group at the Max Planck Institute of Molecular Plant Physiology. This article is part of a project funded by the German Ministry of Research (Plant 2030 Grant agreement number 031B0538) and part of a project that has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (Grant agreement number 810131).
Funding
Open access funding provided by Max Planck Society.
Author information
Authors and Affiliations
Contributions
F.K. suggested the scheme and raised funding; F.K., F.M. and L.Y. suggested experiments; L.Y., S.W., F.M., and E.S. performed the experiments, and supported by F.K. analyzed the data; L.Y. and F.M. wrote the manuscript with F.K. All authors discussed the results and contributed to data interpretation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Takashi Okamoto and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cas9 and gNIA1 transcript presence in grafted wild-type scions.
Tissue samples from transgenic roots (R) and wild-type silique (Sil), flower (Flo), cauline leaf (Cau), stem (St), rosette (Ro) of grafted plants were pooled for RT-PCR detection of Cas9-TLS and gNIA1-TLS fusion transcripts. For each graft combination three independent replicates each representing a pool of three grafted plants are shown which were tested by RT-PCR (45 PCR cycles) for the presence of fusion transcripts. Kanamycin (Kan) transcript expressed only in grafted transgenic rootstocks was used as contamination control in RT-PCR assays. Note that transcript absence of Kan was additionally confirmed by 50 PCR cycles. M: indicates molecular weight markers.
Extended Data Fig. 2 Root expressed mobile Cas9 and gNIA1 induce NIA1 gene editing in the wild-type scion tissues of adult Arabidopsis.
Genotyping PCR to detect NIA1 edited fragments in siliques (Sil) and flowers (Flo). Genomic samples were from the same plant material as analyzed and shown in Fig. 3b. Expected wild-type NIA1 fragment size is 1469 bp. Expected edited NIA1 deletion fragment is approximately 430 bp. Kanamycin (Kan) gene-specific PCRs were used as a control confirming that the detected NIA1 edits were not a result of contamination of wild-type tissues during harvesting. M: indicates molecular weight markers.
Extended Data Fig. 3 Identifying gene edits in the progeny of Col-0 / Cas9 × gNIA1 grafted plants.
Genomic PCR to detect NIA1 edited (deletion) fragments in the wild-type progeny of grafted plants. The smaller PCR amplicons are caused by mobile Cas9 and gRNA - induced deletions in the NIA1 gene. All samples were harvested from 10 days-old seedlings grown NH4 -depleted medium. Each pool contains genomic DNA of ~70 to ~100 seedlings (see Supplementary Table 1). To increase the detection sensitivity of the PCR all pooled samples - except Col-0 / Cas9 x gNIA1 control samples shown in the upper left - were digested by HindIII which targets the wild-type NIA1 sequence but not the edited NIA1 deletion sequence. Wild-type NIA1 fragment size is 1469 bp and the edited deletion fragment is approximately 430 bp. Line # indicates individual grafted plants (lines) from which seeds were collected as pools (P) and analyzed as seedlings by genomic PCR. P # indicates pool of progeny seedlings deriving from a given grafted plant (see also Supplementary Table 1). Note that presence of edits (deletions) was confirmed by additional PCR assays and in part by Sanger sequencing are indicated in red (Fig. 4c).
Extended Data Fig. 4 Gene edits detected in the progeny of wild-type scions grafted on Cas9 x gVenus rootstocks.
a, Schematic diagram showing the 35 Spromoter::H2B-Venus::35Sterminator::Basta cassette targeted by gVenus gRNAs (target1 and 2) and the predicted and PCR detected deletions. The black arrows indicate predicted and the red arrows indicate the detected PCR amplicons created by gVenus target 1 and gVenus target 2 gRNA constructs. Note that the predicted deletion edits were planned in such to remove the H2B-Venus sequence resulting in Basta resistance gene expression (see Supplementary Data 1d). However, the detected deletion edit eliminated part of the 35 S promoter sequence resulting in no expression of the resistance gene. b, Genome edits confirmed by Sanger sequencing. Red arrow indicates detected gRNA 1 mediated editing side which was not predicted, Light blue colored letters indicate the Venus start codon, Black arrows indicate used PCR primers to detected deletion edits. c, Confocal microscopy images of a 6-day-old F1 seedling harvested from a grafted 35Spromoter::H2B-Venus::35Sterminator::Basta / Cas9-TLS2 x gVenus-TLS2 parent showing no Venus fluorescence. This indicates homozygous elimination of the H2B-Venus sequence by deletion editing which was confirmed by genomic PCR assays (right panel). Scale Bar, 350 μm. M: indicates molecular weight markers. d, Editing efficiency analysis of Cas9-TLS / gVenus-TLS constructs in the progeny of grafted plants. # indicates the total number of grafted plants from which the progeny was tested (for details see Supplementary Table 2). * indicates the number of homozygotes identified by loss of Venus fluorescence per 1000 seedlings.
Extended Data Fig. 5 Identifying gene edits in the progeny of Col-0 / Cas9 × gVenus grafted plants.
Genomic PCR to detect gVenus edited (deletion) fragments in the wild-type progeny of grafted plants. The smaller PCR amplicons are caused by mobile Cas9 and gVenus - induced deletions in the Venus gene. All samples were harvested from 10 days-old seedlings grown 0.5 MS medium. Each pool contains genomic DNA of about 40 seedlings (see Supplementary Table 2). To increase the detection sensitivity of the PCR all pooled samples - except Col-0 / Cas9 x gVenus control samples were digested by PstI which targets the wild-type Venus sequence but not the edited Venus deletion sequence. Wild-type Venus fragment size is 1719 bp and the edited deletion fragment is approximately 250 bp. Line # indicates individual grafted plants (lines) from which seeds were collected as pools (P) and analyzed as seedlings by genomic PCR. P # indicates pool of progeny seedlings deriving from a given grafted plant (see also Supplementary Table 2). Note that presence of edits (deletions) confirmed by Sanger sequencing are indicated in red letters. Black arrows indicate detected size of Venus edited bands of approx. 250 bp. Green arrows indicate Venus wild-type expected bands of 1719 bp. M: indicates molecular weight markers.
Extended Data Fig. 6 Cas9 and gNIA1 transcript presence in grafted Brassica rapa / Arabidopsis wild-type scions.
Tissue samples from Arabidopsis transgenic roots (R) and Brassica rapa wild-type silique (Sil), flower (Flo), leaf, stem (St) of grafted plants were used for RT-PCR detection of Cas9-TLS2 and gNIA1-TLS2 fusion transcripts. For each graft combination three independent replicates (each sample was a pool of two grafted plants) are presented which were tested by RT-PCR (45 PCR cycles) for the presence of fusion transcripts. Kanamycin (Kan) transcript expressed only in grafted transgenic rootstocks was used as contamination control in RT-PCR assays. Note that Kan transcript absence was additionally confirmed by 50 PCR cycles. M: indicates molecular weight markers.
Extended Data Fig. 7 Root expressed mobile Cas9 and gNIA1 induce NIA1 gene editing in the wild-type scion tissues of adult Brassica rapa.
Genotyping PCR to detect NIA1 edited fragments in siliques (Sil) and flowers (Flo). Genomic samples were from the same plant material as analyzed and shown in Fig. 5b, c, and Extended Data Fig. 6. The expected wild-type NIA1 fragment size is 1469 bp. The expected edited NIA1 deletion fragment is approximately 430 bp. Kanamycin (Kan) gene-specific PCRs were used as controls confirming that the detected NIA1 edits were not a result of contamination of wild-type tissues during harvesting. M: indicates molecular weight markers.
Extended Data Fig. 8 Schematic diagram showing the construction of gNIA1 expression cassette.
Schematic diagram showing the construction of gNIA1 expression cassette.
Supplementary information
Supplementary Information
Supplementary Tables 1–5 and Supplementary Data 1.
Source data
Source Data Fig. 2
Unprocessed gels.
Source Data Fig. 3
Unprocessed gels.
Source Data Fig. 4
Unprocessed gels.
Source Data Fig. 5
Unprocessed gels.
Source Data Extended Data Fig. 1
Unprocessed gels.
Source Data Extended Data Fig. 2
Unprocessed gels.
Source Data Extended Data Fig. 3
Unprocessed gels.
Source Data Extended Data Fig. 4
Unprocessed gels.
Source Data Extended Data Fig. 5
Unprocessed gels.
Source Data Extended Data Fig. 6
Unprocessed gels.
Source Data Extended Data Fig. 7
Unprocessed gels.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, L., Machin, F., Wang, S. et al. Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks. Nat Biotechnol 41, 958–967 (2023). https://doi.org/10.1038/s41587-022-01585-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-022-01585-8
This article is cited by
-
Arabidopsis cyclophilins direct intracellular transport of mobile mRNA via organelle hitchhiking
Nature Plants (2024)
-
Targeted genome-modification tools and their advanced applications in crop breeding
Nature Reviews Genetics (2024)
-
CRISPR/Cas genome editing in plants: mechanisms, applications, and overcoming bottlenecks
Functional & Integrative Genomics (2024)
-
Plants and global warming: challenges and strategies for a warming world
Plant Cell Reports (2024)
-
Charting the course of plant virology: innovations in diagnostics and beyond—reports from the DPG meeting
Journal of Plant Diseases and Protection (2024)