Abstract
The influence of genetic background on driver mutations is well established; however, the mechanisms by which the background interacts with Mendelian loci remain unclear. We performed a systematic secondary-variant burden analysis of two independent cohorts of patients with Bardet–Biedl syndrome (BBS) with known recessive biallelic pathogenic mutations in one of 17 BBS genes for each individual. We observed a significant enrichment of trans-acting rare nonsynonymous secondary variants in patients with BBS compared with either population controls or a cohort of individuals with a non-BBS diagnosis and recessive variants in the same gene set. Strikingly, we found a significant over-representation of secondary alleles in chaperonin-encoding genes—a finding corroborated by the observation of epistatic interactions involving this complex in vivo. These data indicate a complex genetic architecture for BBS that informs the biological properties of disease modules and presents a model for secondary-variant burden analysis in recessive disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
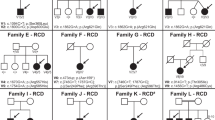
A summary of the genotypes presented in the present study is given in Supplementary Table 1. However, restrictions apply to the availability of whole-exome data, patient consent forms and Institutional Review Board instructions. Selected data can be made available from the corresponding author upon reasonable request and inclusion of appropriate Institutional Review Board documentation.
References
Skafidas, E. et al. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol. Psychiatry 19, 504–510 (2014).
Beales, P. L. et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet–Biedl syndrome. Am. J. Hum. Genet. 72, 1187–1199 (2003).
Katsanis, N. et al. Triallelic inheritance in Bardet–Biedl syndrome, a Mendelian recessive disorder. Science 293, 2256–2259 (2001).
Badano, J. L. et al. Dissection of epistasis in oligogenic Bardet–Biedl syndrome. Nature 439, 326–330 (2006).
Cardenas-Rodriguez, M. et al. The Bardet–Biedl syndrome-related protein CCDC28B modulates mTORC2 function and interacts with SIN1 to control cilia length independently of the mTOR complex. Hum. Mol. Genet. 22, 4031–4042 (2013).
Shaheen, R. et al. Characterizing the morbid genome of ciliopathies. Genome Biol. 17, 242 (2016).
Otto, E. A. et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 42, 840–850 (2010).
Li, B. & Leal, S. M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83, 311–321 (2008).
Lin, M. F. et al. Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. Genome Res. 21, 1916–1928 (2011).
Henikoff, S. & Henikoff, J. G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA 89, 10915–10919 (1992).
Loktev, A. V. et al. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev. Cell 15, 854–865 (2008).
Nachury, M. V. et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 (2007).
Wei, Q. et al. The BBSome controls IFT assembly and turnaround in cilia. Nat. Cell Biol. 14, 950–957 (2012).
Garcia-Gonzalo, F. R. et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43, 776–784 (2011).
Seeger-Nukpezah, T. et al. The centrosomal kinase Plk1 localizes to the transition zone of primary cilia and induces phosphorylation of nephrocystin-1. PLoS ONE 7, e38838 (2012).
Kim, J. C. et al. MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet–Biedl syndrome, is a novel centrosomal component required for cytokinesis. J. Cell Sci. 118, 1007–1020 (2005).
Stoetzel, C. et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet–Biedl syndrome. Am. J. Hum. Genet. 80, 1–11 (2007).
Leitch, C. C. et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet–Biedl syndrome. Nat. Genet. 40, 443–448 (2008).
Lindstrand, A. et al. Recurrent CNVs and SNVs at the NPHP1 locus contribute pathogenic alleles to Bardet–Biedl syndrome. Am. J. Hum. Genet. 94, 745–754 (2014).
Lindstrand, A. et al. Copy-number variation contributes to the mutational load of Bardet–Biedl syndrome. Am. J. Hum. Genet. 99, 318–336 (2016).
Stoetzel, C. et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat. Genet. 38, 521–524 (2006).
Zaghloul, N. A. et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet–Biedl syndrome. Proc. Natl Acad. Sci. USA 107, 10602–10607 (2010).
Rutherford, S. L. & Lindquist, S. Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342 (1998).
Karras, G. I. et al. HSP90 shapes the consequences of human genetic variation. Cell 168, 856–866.e12 (2017).
Gonzaga-Jauregui, C. et al. Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep. 12, 1169–1183 (2015).
Nikopoulos, K. et al. A frequent variant in the Japanese population determines quasi-Mendelian inheritance of rare retinal ciliopathy. Nat. Commun. 10, 2884 (2019).
Beales, P. L., Elcioglu, N., Woolf, A. S., Parker, D. & Flinter, F. A. New criteria for improved diagnosis of Bardet–Biedl syndrome: results of a population survey. J. Med. Genet. 36, 437–446 (1999).
Hjeij, R. et al. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 93, 357–367 (2013).
Redin, C. et al. Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet–Biedl and Alström syndromes. J. Med. Genet. 49, 502–512 (2012).
Yang, Y. et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N. Engl. J. Med. 369, 1502–1511 (2013).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Price, A. L. et al. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86, 832–838 (2010).
Zhan, X., Hu, Y., Li, B., Abecasis, G. R. & Liu, D. J. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 32, 1423–1426 (2016).
Sealfon, R. S. et al. FRESCo: finding regions of excess synonymous constraint in diverse viruses. Genome Biol. 16, 38 (2015).
Lindblad-Toh, K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011).
Acknowledgements
We thank the patients and their families for participating in this study, as well as their caring physicians who provided clinical information and referred the patients for diagnostic analyses. We are grateful to C. Glodosky and A. Krentz from PreventionGenetics and R. Haws from Marshfield Clinic for providing information on clinical BBS exome data. We thank A.-S. Jaeger and M. Antin for technical assistance. This study was supported by NIH grants GM121317-13, HD042601 and DK072301 (to N.K.), grants R35GM127131, R01MH101244 and U01HG00908 (to S.R.S.) and R01 HG004037 (to I.J.), as well as GENCODE Wellcome Trust grant U41 HG007234 (to I.J.). R.A.L. is a senior scientific investigator of research to prevent blindness. N.K. is a distinguished Valerie and George D. Kennedy professor.
Author information
Authors and Affiliations
Contributions
M. Kousi, E.E.D., S.R.S. and N.K. designed the study. M. Kousi, A.O. and K.M. performed the experimental work by genotyping and analyzing the sequencing data. O.S., N.M., S.A., C.A.C., M. Kousi, H.B., M.E.T. and S.R.S. performed the statistical analyses assessing mutational burden and population structure. I.J., M.Y.W. and M. Kellis performed the analyses assessing the impact of the identified changes on genomic sequence signatures. J.M., J.A.M., R.A.L. and H.D. contributed the samples that were analyzed in this study. A.S. collated the genetic information of the discovery cohort. M. Kousi performed the collective analysis of genetic data and modular interactions. N.K. and M. Kousi wrote the manuscript. All authors read and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
N.K. is a founder of, and holds significant stock in, Rescindo Therapeutics.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
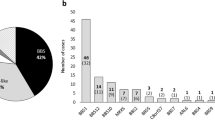
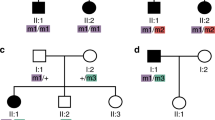
Extended Data Fig. 1 Distribution of burden-contributing variation across case and control cohorts.
a, Burden-contributing variants across cases of the Discovery cohort (n = 102), Replication cohort (n = 175), and Meta-analysis of all BBS cases (n = 277) across four population-based minor allele frequency (MAF) cutoffs (1%, 0.5%, 0.1%, and 0.001%). b, Values of burden-contributing variation between BBS cases of the Discovery cohort (n = 102) and the cohort of NEU controls (n = 384) showing a 2.5-fold enrichment for ultra-rare (MAF < 0.001%) alleles in cases compared to controls. c, BBS cases of the Replication cohort (n = 175) and the Replication control cohort (n = 488) showing a 2.5-fold enrichment for ultra-rare (MAF < 0.001%) alleles in cases compared to controls. d, Collapsed case, control and “non-BBS recessive” cohorts. e, f, Distribution of individuals with burden-contributing alleles with MAF < 1% (e) and with MAF < 0.001% (f) in control individuals (blue bars) and BBS cases (orange bars).
Extended Data Fig. 2 Distribution of burden-contributing variation across case and control cohorts in each of four discrete MAF bins.
a, The Discovery case cohort shows a 2.5-fold enrichment for ultra-rare (0.001% > MAF > 0%) alleles compared to controls. b, The Replication cohort shows a 2-fold enrichment of such alleles compared to the exome control cohort. c, The BBS case meta-analysis shows a 2.2-fold enrichment compared to the combined control cohorts. d, Collapsed cohorts. a–d show plots across four MAF bins (1% > MAF > 0.5%, 0.5% > MAF > 0.1%, 0.1% > MAF > 0.001%, and 0.001% > MAF > 0%).
Extended Data Fig. 3 Estimate of protein impact for the least disruptive of the diagnostic variants for BBS cases and non-BBS recessive individuals, with at least one missense change in the primary locus.
With high BLOSUM62 scores denoting biochemically similar amino acid changes and lower scores marking radical amino acid changes, the graph shows evidence for bona fide BBS cases harboring more disruptive variants.
Supplementary information
Supplementary Information
Supplementary Tables 2–8
Supplementary Table
Supplementary Table 1
Rights and permissions
About this article
Cite this article
Kousi, M., Söylemez, O., Ozanturk, A. et al. Evidence for secondary-variant genetic burden and non-random distribution across biological modules in a recessive ciliopathy. Nat Genet 52, 1145–1150 (2020). https://doi.org/10.1038/s41588-020-0707-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-0707-1
This article is cited by
-
Skeletal ciliopathy: pathogenesis and related signaling pathways
Molecular and Cellular Biochemistry (2024)
-
Lethal neonatal respiratory failure due to biallelic variants in BBS1 and monoallelic variant in TTC21B
Pediatric Nephrology (2023)
-
Secondary-variant genetic burden in Bardet–Biedl syndrome
Nature Reviews Nephrology (2021)