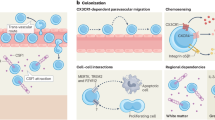
Abstract
Microglia are the resident macrophages of the CNS that serve critical roles in brain construction. Although human brains contain microglia by 4 weeks gestation, an understanding of the earliest microglia that seed the brain during its development remains unresolved. Using time-lapse imaging in zebrafish, we discovered a mrc1a+ microglia precursor population that seeds the brain before traditionally described microglia. These early microglia precursors are dependent on lymphatic vasculature that surrounds the brain and are independent of pu1+ yolk sac-derived microglia. Single-cell RNA-sequencing datasets reveal Mrc1+ microglia in the embryonic brains of mice and humans. We then show in zebrafish that these early mrc1a+ microglia precursors preferentially expand during pathophysiological states in development. Taken together, our results identify a critical role of lymphatics in the microglia precursors that seed the early embryonic brain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data collected for the study are included in the figures. For all datasets that did not pass normality tests, secondary unpaired non-parametric t-test analyses were used and yielded similar P values. Source data are provided with this paper.
Code availability
All code for the scRNA-seq data analysis can be accessed at https://github.com/michael-r-odea/Green_ODea_2022/.
References
Miyamoto, A. et al. Microglia contact induces synapse formation in developing somatosensory cortex. Nat. Commun. 7, 12540 (2016).
Schafer, D. et al. Microglia sculpt postnatal neuronal circuits in an activivty and complement-dependent manner. Neuron 74, 691–705 (2012).
Paolicelli, R. C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458 (2011).
Cunningham, C. L., Martinez-Cerdeno, V. & Noctor, S. C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 33, 4216–4233 (2013).
Marín-Teva, J. L. et al. Microglia promote the death of developing Purkinje cells. Neuron 41, 535–547 (2004).
Wakselman, S. et al. Developmental neuronal death in hippocampus requires the microglial CD11b integrin and DAP12 immunoreceptor. J. Neurosci. 28, 8138–8143 (2008).
Ueno, M. et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 16, 543–551 (2013).
Shigemoto-Mogami, Y., Hoshikawa, K., Goldman, J. E., Sekino, Y. & Sato, K. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci. 34, 2231–2243 (2014).
Hughes, A. N. & Appel, B. Microglia phagocytose myelin sheaths to modify developmental myelination. Nat. Neurosci. https://doi.org/10.1038/s41593-020-0654-2 (2020).
Choi, B. H. Hematogenous cells in the central nervous system of developing human embryos and fetuses. J. Comp. Neurol. 196, 683–694 (1981).
Andjelkovic, A. V., Nikolic, B., Pachter, J. S. & Zecevic, N. Macrophages/microglial cells in human central nervous system during development: an immunohistochemical study. Brain Res. 814, 13–25 (1998).
Monier, A. et al. Entry and distribution of microglial cells in human embryonic and fetal cerebral cortex. J. Neuropathol. Exp. Neurol. 66, 372–382 (2007).
Verney, C., Monier, A., Fallet-Bianco, C. & Gressens, P. Early microglial colonization of the human forebrain and possible involvement in periventricular white-matter injury of preterm infants. J. Anat. 217, 436–448 (2010).
Menassa, D. A. & Gomez-Nicola, D. Microglial dynamics during human brain development. Front. Immunol. 9, 1014 (2018).
Monier, A., Evrard, P., Gressens, P. & Verney, C. Distribution and differentiation of microglia in the human encephalon during the first two trimesters of gestation. J. Comp. Neurol. 499, 565–582 (2006).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Herbom, P., Thisse, B. & Thisse, C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina and epiderm is through a M-CSF receptor-dependent invasive process. Dev. Biol. 288, 274–288 (2001).
Casano, A. M., Albert, M. & Peri, F. Developmental apoptosis mediates entry and positioning of microglia in the zebrafish brain. Cell Rep. 16, 897–906 (2016).
Xu, J. et al. Temporal-spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev. Cell 34, 632–641 (2015).
Ferrero, G. et al. Embryonic microglia derive from primitive macrophages and are replaced by cmyb-dependent definitive microglia in zebrafish. Cell Rep. 24, 130–141 (2018).
Chen, S. K. et al. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell 141, 775–785 (2010).
Chen, H. et al. Fate mapping via CCR2-CreER mice reveals monocyte-to-microglia transition in development and neonatal stroke. Sci. Adv. 6, eabb2119 (2020).
Ohnmacht, J. et al. Spinal motor neurons are regenerated after mechanical lesion and genetic ablation in larval zebrafish. Development 143, 1464–1474 (2016).
Peri, F. & Nüsslein-Volhard, C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell 133, 916–927 (2008).
Jung, H. M. et al. Development of the larval lymphatic system in zebrafish. Development 144, 2070–2081 (2017).
van Lessen, M. et al. Intracellular uptake of macromolecules by brain lymphatic endothelial cells during zebrafish embryonic development. Elife 6, 1–24 (2017).
Galanternik, M. V. et al. A novel perivascular cell population in the zebrafish brain. Elife 6, 1–28 (2017).
Shin, M. et al. Vegfc acts through ERK to induce sprouting and differentiation of trunk lymphatic progenitors. Development 144, 531 (2017).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell–state changes. Immunity 50, 253–271 (2018).
Plein, A., Fantin, A., Denti, L., Pollard, J. W. & Ruhrberg, C. Erythro-myeloid progenitors contribute endothelial cells to blood vessels. Nature 562, 223–228 (2018).
Koltowska, K. et al. Vegfc regulates bipotential precursor division and Prox1 expression to promote lymphatic identity in zebrafish. Cell Rep. 13, 1828–1841 (2015).
Green, L. A., Nebiolo, J. C. & Smith, C. J. Microglia exit the CNS in spinal root avulsion. PLoS Biol. 17, e3000159 (2019).
Kracht, L. et al. Human fetal microglia acquire homeostatic immune-sensing properties early in development. Science 369, 530–537 (2020).
Utz, S. G. et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell 181, 557–573 (2020).
Kierdorf, K. et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013).
Bennett, M. L. et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl Acad. Sci. USA 113, E1738–E1746 (2016).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Herbomel, P., Thisse, B. & Thisse, C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745 (1999).
Goldmann, T. et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805 (2016).
Smith, C. J., Morris, A. D., Welsh, T. G. & Kucenas, S. Contact-mediated inhibition between oligodendrocyte progenitor cells and motor exit point glia establishes the spinal cord transition zone. PLoS Biol. 12, e1001961 (2014).
Astin, J. W. et al. An in vivo antilymphatic screen in zebrafish identifies novel inhibitors of mammalian lymphangiogenesis and lymphatic-mediated metastasis. Mol. Cancer Ther. 13, 2450–2462 (2014).
Zhang, L. et al. VEGFR-3 ligand-binding and kinase activity are required for lymphangiogenesis but not for angiogenesis. Cell Res. 20, 1319–1331 (2010).
Tammela, T. et al. Photodynamic ablation of lymphatic vessels and intralymphatic cancer cells prevents metastasis. Sci. Transl. Med. 3, 69ra11 (2011).
Kilarski, W. W. et al. Optimization and regeneration kinetics of lymphatic-specific photodynamic therapy in the mouse dermis. Angiogenesis 17, 347–357 (2014).
Elmore, M. R. P. et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014).
Erblich, B., Zhu, L., Etgen, A. M., Dobrenis, K. & Pollard, J. W. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS ONE 6, e26317 (2011).
Li, Q. & Barres, B. A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. https://doi.org/10.1038/nri.2017.125 (2017).
Smith, C. J., Johnson, K., Welsh, T. G., Barresi, M. J. F. & Kucenas, S. Radial glia inhibit peripheral glial infiltration into the spinal cord at motor exit point transition zones. Glia 64, 1138–1153 (2016).
Cuadros, M. A., Martin, C., Coltey, P., Almendros, A. & Navascués, J. First appearance, distribution and origin of macrophages in the early development of the avian central nervous system. J. Comp. Neurol. 330, 113–129 (1993).
Gritz, E. & Hirschi, K. K. Specification and function of hemogenic endothelium during embryogenesis. Cell. Mol. Life Sci. 73, 1547–1567 (2016).
Lancrin, C. et al. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457, 892–895 (2009).
Swiers, G. et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 4, 2924 (2013).
Boisset, J. C. et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010).
Jaffredo, T., Gautier, R., Eichmann, A. & Dieterlen-Lièvre, F. Intra-aortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 125, 4575–4583 (1998).
Jordan, H. E. Evidence of hemogenic capacity of endothelium. Anat. Rec. 10, 417–420 (1916).
Nakano, H. et al. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat. Commun. 4, 1–10 (2013).
Li, Z. et al. Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell 11, 663–675 (2012).
Cugurra, A. et al. Skull and vertebral bone marrow are myeloid reservoirs for the meninges and CNS parenchyma. Science 373, eabf7844 (2021).
Da Mesquita, S. et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature 560, 185–191 (2018).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015).
Castranova, D. et al. Live imaging of intracranial lymphatics in the zebrafish. Circ. Res. 128, 42–58 (2021).
Sieger, D., Moritz, C., Ziegenhals, T., Prykhozhij, S. & Peri, F. Long-range Ca2+ waves transmit brain-damage signals to microglia. Dev. Cell 22, 1138–1148 (2012).
Kucenas, S., Wang, W.-D., Knapik, E. W. & Appel, B. A selective glial barrier at motor axon exit points prevents oligodendrocyte migration from the spinal cord. J. Neurosci. 29, 15187–15194 (2009).
Kirby, B. B. et al. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat. Neurosci. 9, 1506–1511 (2006).
Andermann, P., Ungos, J. & Raible, D. W. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev. Biol. 251, 45–58 (2002).
McGraw, H. F., Snelson, C. D., Prendergast, A., Suli, A. & Raible, D. W. Postembryonic neuronal addition in zebrafish dorsal root ganglia is regulated by Notch signaling. Neural Dev. 7, 23 (2012).
Langenau, D. M. et al. In vivo tracking of T cell development, ablation and engraftment in transgenic zebrafish. Proc. Natl Acad. Sci. USA 101, 7369–7374 (2004).
Lawson, N. D. & Weinstein, B. M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 (2002).
Rosenberg, A. F., Wolman, M. A., Franzini-Armstrong, C. & Granato, M. In vivo nerve–macrophage interactions following peripheral nerve injury. J. Neurosci. 32, 3898–3909 (2012).
Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Nichols, E. L., Green, L. A. & Smith, C. J. Ensheathing cells utilize dynamic tiling of neuronal somas in development and injury as early as neuronal differentiation. Neural Dev. 13, 19 (2018).
Nichols, E. L. & Smith, C. J. Pioneer axons employ Cajal’s battering ram to enter the spinal cord. Nat. Commun. 10, 562 (2019).
Kikel-Coury N. L. et al. Identification of astroglia-like cardiac nexus glia that are critical regulators of cardiac development and function. PLoS Biol. 19, e3001444 (2021).
Kikel-Coury, N. L. et al. Pioneer axons utilize a Dcc signaling-mediated invasion brake to precisely complete their pathfinding odyssey. J. Neurosci. 41, 6617–6636 (2021).
R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.r-project.org/ (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Zou, Z., Hua, K. & Zhang, X. HGC: fast hierarchical clustering for large-scale single-cell data. Bioinformatics 37, btab420 (2021).
Krueger, F., James, F., Ewels, P., Afyounian, E. & Schuster-Boeckler, B. FelixKrueger/TrimGalore: v0.6.7. https://doi.org/10.5281/zenodo.5127899 (2021).
Patro, R et al. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods https://doi.org/10.1038/nmeth.4197 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics https://doi.org/10.1093/bioinformatics/btw313 (2016).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Rainer, J. EnsDb.Hsapiens.v79: Ensembl-based annotation package. R package version 2.99.0. https://bioconductor.org/packages/release/data/annotation/html/EnsDb.Hsapiens.v79.html (2017).
Durinck, S., Spellman, P. T., Birney, E., Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. https://doi.org/10.1038/nprot.2009.97 (2009).
Wickham H. ggplot2: elegant Graphics for Data Analysis (Springer International Publishing, 2016) https://doi.org/10.1007/978-3-319-24277-4
Kassambara, A. ggpubr: ‘ggplot2’-based publication-ready plots. CRAN R-project https://rdrr.io/cran/ggpubr/ (2019).
Nichols, E. L. & Smith, C. J. Synaptic-like vesicles facilitate pioneer axon invasion. Curr Biol. https://doi.org/10.1016/j.cub.2019.06.078 (2019).
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction Kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007).
Prendergast, A. et al. The metalloproteinase inhibitor Reck is essential for zebrafish DRG development. Development 139, 1141–1152 (2012).
Green, L. & Smith, C. J. Single-cell photoconversion in living intact zebrafish. J. Vis. Exp. https://doi.org/10.3791/57024 (2018).
Hoshijima, K. et al. Highly efficient CRISPR–Cas9-based methods for generating deletion mutations and F0 embryos that lack gene function in zebrafish. Dev. Cell 51, 645–657 (2019).
Johnson, K. et al. Gfap-positive radial glial cells are an essential progenitor population for later-born neurons and glia in the zebrafish spinal cord. Glia 64, 1170–1189 (2016).
Acknowledgements
We thank B. Weinstein (National Institutes of Health (NIH)) for sending us the Tg(mrc1a:egfp) animals, D. Seiger for p5e-pu1, and W. Clements for Tg(lck:gfp) and Tg(fli1:gfp) animals. We thank B. Stevens, T. Hammond, K. Monk, C. Bennett and S. Zhang for their helpful comments and reagent guidance. We also thank B. Redford, S. Connell and 3i for imaging-related questions, S. Cole in the NDiiF Optical Microscopy Core for help with light-sheet imaging (OMC/NDIIF and the National Science Foundation-Major Research Instrumentation Program 1919832) and IMARIS analysis, and D. Bang, K. Heed and B. Gervais for zebrafish housing and upkeep. This work was supported by the University of Notre Dame, the Elizabeth and Michael Gallagher Family, Centers for Zebrafish Research and Stem Cells and Regenerative Medicine at the University of Notre Dame, the Indiana Spinal Cord and Brain Injury Research with the Indiana State Board of Health (C.J.S.), the Alfred P. Sloan Foundation (C.J.S.) and the NIH (DP2NS117177; C.J.S.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
L.A.G., M.R.O. and C.A.H. performed the analysis, experimentation, writing and editing of the manuscript. D.F.D. performed experimentation. M.R.O., L.A.G. and C.J.S. conceived the study. C.J.S. wrote and edited the manuscript and supervised and funded the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Antoine Louveau and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
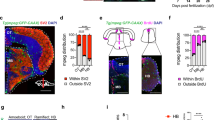
Extended Data Fig. 1 mrc1a+ microglia are distinct from other BLEC and vascular endothelial cells.
(A) Quantification of the percentage of pu1+;4C4+ microglia in Tg(pu1:Gal4;UAS:rfp) compared to Tg(pu1:eos) animals (t-test: Tg(pu1:gal4-uas:tagrfp) vs Tg(pu1:eos) p = 0.1838; two-tailed) (n = 40 animals). (B) Quantification of the average number of mrc1a+ only cells in Tg(mrc1a:egfp);Tg(nbt:dsred) animals from 2 dpf − 5 dpf (n = 20 animals). (C) Quantification of the average number of 4C4+ only cells in Tg(mrc1a:egfp);Tg(nbt:dsred) animals from 2 dpf − 5 dpf (n = 20 animals). (D) Quantification of the average number of mrc1a+;4C4+ cells in Tg(mrc1a:egfp);Tg(nbt:dsred) animals from 2 dpf − 5 dpf (n = 20 animals). (E) Quantification of the average number of microglia with marked expression in Tg(mrc1a:egfp);Tg(gfap:nsfb-mCherry) and Tg(mrc1a:egfp);Tg(nbt:dsred) animals at 2 dpf and 5 dpf (t-test: 2 dpf Tg(mrc1a:egfp);Tg(gfap:nsfb-mcherry) versus Tg(mrc1a:egfp);Tg(nbt:dsred) p = 0.0428, 5 dpf Tg(mrc1a:egfp);Tg(gfap:nsfb-mcherry) vs. Tg(mrc1a:egfp);Tg(nbt:dsred) p = 0.0738; all two-tailed)(n = 28 animals). (F) Confocal z-projection images of Tg(mrc1a:egfp) animals at 5 dpf stained with 4C4, Lcp1, Prox1, and Flt4. White arrowheads indicate 4C4+ microglia. Orange arrowheads indicate a small number of 4C4+;Lcp1+ microglia. Purple arrowheads indicate Flt4+ cells located along the mrc1a+ vessel endothelium that are not 4C4+. (G) Quantification of the average number of mrc1a+ parenchyma cells with mrc1a+ only expression compared to mrc1a+;apoeb+ microglia (n = 7 animals). Imaging window represents one 0.0027 mm3 region per animal (A-G). Scale bar equals 10 µm (B).
Extended Data Fig. 2 mrc1a+ microglia function like traditional microglia.
(A) Confocal z-projection images of 5 dpf Tg(mrc1a:egfp) animals stained with antibodies or other transgenic animals to label debris from synaptic, neuronal, oligodendrocyte, microglia, or astroglia populations. White boxes indicate regions of engulfed debris puncta. Arrows indicate individual debris puncta within mrc1a+ microglia. (B) Confocal z-projection images of 5 dpf Tg(pu1:Gal4;UAS:gfp) animals stained with same antibodies /transgenic animals represented in (A). Arrows indicate individual debris puncta within mrc1a+ microglia. (C) Confocal z-projection still images from a 24 hour timelapse of Tg(mrc1a:egfp);Tg(pu1:Gal4;UAS:rfp) animals from 4 dpf to 5 dpf showing homotypic interactions between mrc1a+ and mrc1a+ microglia and pu1+ and pu1+ microglia populations. White arrowheads indicate mrc1a+ microglia and blue arrowheads indicate pu1+ microglia. Dashed yellow box indicates contact point for two microglia. White arrowheads indicate mrc1a+ microglia. Blue arrowheads indicate pu1+ microglia. (D) Quantification of the migration path two individual mrc1a+ microglia traveled pre and post contact (n = 7 animals). (E) Quantification of the migration path two individual pu1+ microglia traveled pre and post contact (n = 7 animals). Imaging window equals 0.0027 mm3 (A,B), 0.0081 mm3 (C-E). Scale bar equals 10 µm (A,B), 100 µm (C).
Extended Data Fig. 3 Mrc1 is expressed in developmental microglia in the mammalian brain.
Panels A-D refer to analysis of data from Hammond et al. (2019)36. (A) UMAP of initial clustering of all cells from E14, P4 & P5, and P30 from Hammond et al (2019). Clusters A-G represent microglia; cluster H is macrophages/monocytes; cluster I is endothelial cells; and clusters J and K are neuronal. (B) Violin plot of log-normalized expression of Mrc1, microglia markers (Tmem119, P2ry12), macrophage/monocyte markers (F13a1, Ccr1, Ccr2), endothelial markers (Cldn5, Vtn, Pecam1), and neuronal markers (Neurod6, Nfib, Elavl3). Clusters A-H were chosen for subclustering to identify microglial subpopulations. (C) Heatmap of z-scored average expression of the top 10 differentially expressed genes for each of the 16 clusters identified in the second round of clustering of microglia and macrophages. (D) Violin plot comparing expression of Spi1 (the gene encoding the PU.1 transcription factor) in Mrc1+ and Mrc1- microglia (both aggregated from clusters 1 & 4-16). Panels E-H refer to analysis of data from Kracht et al., (2020)41. (E) UMAP of initial clustering of all cells from Kracht et al. (2020). Clusters A-D are microglia; cluster E is monocytes/macrophages; cluster F is neurons; and cluster G is erythrocytes. (F) Violin plots of log-normalized expression of canonical microglia, macrophage/monocyte, neuronal, and erythrocytic genes across the seven initial clusters. Clusters A-E were chosen for subclustering to identify microglia and macrophage subpopulations. (G) Heatmap of z-scored average expression of top 10 differentially expressed genes per cluster across all 10 clusters identified in the sub-clustering of microglia and macrophages from Kracht et al. (2020). (H) Violin plot comparing expression of SPI1 (the gene encoding the PU.1 transcription factor) in MRC1+ and MRC1- microglia (both aggregated from clusters 1 & 4-16). Points were added for single cells as a low proportion of cells in both groups expressed detectable MRC1. For (D) and (H) P-value refers to a Wilcoxon rank-sum test with Bonferroni correction for multiple comparisons. “Avg. log2FC” refers to the average log2-fold change in expression between Mrc1/MRC1+ and Mrc1/MRC1- microglia. Positive values refer to higher expression in Mrc1/MRC1+ microglia. The percentage of cells in each group expressing at least one detected read of Mrc1/MRC1 is shown in parentheses. Statistical significance in differential expression testing was determined by Wilcoxon rank-sum test (ɑ = 0.05, with Bonferroni correction) and an average log fold-change threshold of ±0.5.
Extended Data Fig. 4 mrc1a+ microglia are dependent on lymphangiogenesis.
(A) Representative images of two orthogonal rotations of confocal z-projection (left) and IMARIS 3D surface rendering (right) of 6 dpf Tg(mrc1a:egfp) animals showing a secondary sprout of a growing lymphatic vessel. White arrows indicate vessel secondary sprout site. Blue arrowheads indicate hollow vessel center. (B) Representative confocal z-projections of Tg(mrc1a:egfp) animals stained with 4C4 showing the reduction of mrc1a+;4C4+ microglia in cinnarizine, flunarizine, and leflunomide treated animals compared to DMSO control animals. Blue arrowheads represent mrc1a+;4C4+ microglia. (C) Representative confocal z-projections of 5 dpf Tg(mrc1a:egfp) animals showing the disruption of vessel growth and development in animals treated with A77-1726, cinnarizine, flunarizine, or leflunomide compared to control DMSO animals. (D) Quantification showing the reduced average length of brain lymphatic vessels in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. A77-1726 p = 0.0444, Mean diff=22.94, DF-358, q = 2.622, SE of diff=8.749; DMSO vs. cinnarizine p = 0.2378, Mean diff=16.62, DF = 358, q = 1.937, SE of diff=8.592; DMSO vs. flunarizine p = 0.0060, Mean diff=35.94, DF = 358, q = 3.263, SE of diff=11.02; DMSO vs. leflunomide p = 0.0008, Mean diff=25.83, DF = 358, q = 3.81, SE of diff=6.78) (n = 80 animals) (E) Quantification showing the reduced average number of secondary sprouts in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. A77-1726 p = 0.0207, mean diff =1.8, DF = 63, q = 2.966, SE of diff=0.6068; DMSO vs. cinnarizine p = 0.0012, Mean diff=2.371, DF = 63, q = 3.908, SE of diff=0.6068; DMSO vs. flunarizine p = 0.0178, Mean diff=2.336, DF = 63, q = 3.019, SE of diff=0.7736; DMSO vs. leflunomide p = 0.0004, Mean diff=2.308, DF = 63, q = 4.213, SE of diff=0.5478)(n = 80 animals). (F) Quantification showing the reduced average number of lymphatic vessels surrounding the brain in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. A77-1726, DMSO vs. cinnarizine p = 0.0081, Mean diff=3.242, DF = 64, q = 3.29, SE of diff=0.9854; DMSO vs. cinnarizine p = 0.0124, Mean diff=3.099, DF = 64, q = 3.145, SE of diff=0.9854; DMSO vs. flunarizine p = 0.0883, Mean diff=3.028, DF = 64, q = 2.408, SE of diff=1.257; DMSO vs. leflunomide p = 0.1368, Mean diff=1.972, DF = 64, q = 2.218, SE of diff=0.889)(n = 80 animals). (G) Quantification of the number of 4C4+ only microglia in DMSO control animals compared to leflunomide and flunarizine treated animals (t-test: DMSO vs. leflunomide, DMSO vs. flunarizine (Ordinary one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. leflunamide p = 0.9997, Mean diff = −0.04274, DF = 25, q = 0.01816, SE of diff=2.353; DMSO vs. flunarizine p = 0.2097, Mean diff=4.346, DF = 25, q = 1.623, SE of diff=2.678)(n = 29 animals). (H) Quantification of the number of pu1+ only microglia in DMSO control animals compared to leflunomide and flunarizine treated animals (Ordinary one-way ANOVA/Dunnett’s multiple comparisonst: DMSO vs. leflunamide p = 0.9932, Mean diff = −0.2222, DF = 36, q = 0.09986, SE of diff=2.225; DMSO vs. flunarizine p = 0.6213, Mean diff = −2.167, DF = 25, q = 0.8554, SE of diff=2.533)(n = 29 animals). (I) Quantification of the number of pu1+;4C4+ microglia in DMSO control animals compared to leflunomide and flunarizine treated animals (Ordinary one-way ANOVA/Dunnett’s multiple comparisons: DMSO vs. Leflunamide p = 0.7997, Mean diff = −1.342, DF-25, q = 1.19, SE of diff=1.167; DMSO vs. Flunarizine p = 0.4353, Mean diff=0.7692, DF = 25, q = 0.5789, SE of diff=1.329)(n = 29 animals). Imaging window equals 0.0027 mm3 (A-F), 0.0081 mm3 or 3000 µm (G-I). Scale bar equals 10 µm (A-C).
Extended Data Fig. 5 mrc1a+ microglia are dependent on lymphangiogenesis.
(A) Quantification of the total number of mrc1a+ only microglia in uninjected and Cas9 only animals compared to flt4 gRNA injected animals (one-way ANOVA/Dunnett’s multiple comparisons: uninjected vs. Cas9 only injected p = 0.9389, Mean diff = =0.08359, DF = 18, q = 0.4783, SE of diff=0.2472; Cas9 only injected vs. flt4 gRNA/Cas9 injected p = 0.9377, Mean diff = −0.003759, DF = 118, q = 0.02349, SE of diff=0.2263; uninjected vs. flt4 gRNA/Cas9 injected p = 0.9998, Mean diff=0.07983, DF = 118, q = 0.4831, SE of diff=0.2337)(n = 122 animals). (B) Quantification of the total number of 4C4+ only microglia in uninjected and Cas9 only animals compared to flt4 gRNA injected animals (one-way ANOVA/Dunnett’s multiple comparisons: uninjected vs. Cas9 only injected p = 0.0573, Mean diff = −1.423, DF = 118, q = 3.277, SE of diff=0.614; Cas9 only injected vs. flt4 gRNA/Cas9 injected p = 0.6773, Mean diff = −0.9334, DF = 118, q = 2.348, SE of diff=0.5622; uninjected vs. flt4 gRNA/Cas9 injected p = 0.2249, Mean diff=0.4892, DF = 118, q = 1.192, SE of diff=0.5805)(n = 122 animals). (C) Confocal z-projections of the brain, RBI, and Yolksac regions in Tg(mrc1a:egfp) animals injected with bactin:eos pre and post-photoconversion. Purple arrowheads indicate successfully photoconverted pEos+ cells. (D) Quantification of the average number of mrc1a+ only cells in the brain at 56 hpf following photoconversion of bactin:eos in the brain vessels, RBI, and yolk sac (t-test: bra(n = 17 animals) in vs. RBI p = 0.9467, RBI vs. yolksac p = 0.8988, brain vs. yolksac p = 0.8611; all two-tailed). Imaging window equals 0.0027 mm3 (A-D). Scale bar equals 10 µm (C).
Extended Data Fig. 6 mrc1a+ microglia are distinct from yolk-sac derived microglia.
(A) Quantification of the average number of mrc1a+ only cells in the brain imaging window over time (n = 69 animals). (B) Quantification of the average number of 4C4+ only cells in the brain imaging window over time. (C) Quantification of the average number of pu1+ only cells in the brain imaging window over time (n = 69 animals). (D) Quantification of the average number of mrc1a+;4C4+ cells in the brain imaging window over time (n = 69 animals). (E) Quantification of the average number of mrc1a+;pu1+ cells in the brain imaging window over time (n = 69 animals). (F) Quantification of the average number of mrc1a+;pu1+;4C4+ cells in the brain imaging window over time (n = 69 animals). (G) Quantification of compiled average number of mrc1a+ only, 4C4+ only, pu1+ only, and mrc1a+;4C4+ cells over time. (H) Quantification of the total number of mrc1a+;pu1+ cells per imaging window in uninjected animals compared to spi1b sgRNA injected animals (t-test: uninjected vs. spi1b gRNA/Cas9 injected p = 0.0003’two-tailed)(n = 41 animals). (I) Quantification of the total number of 4C4+ only cells per imaging window in uninjected animals compared to spi1b sgRNA injected animals (t-test: uninjected vs. spi1b/Cas9 injected p = 0.2849;two-tailed)(n = 41 animals). (J) Representative confocal z-projection images of Tg(mrc1a:egfp); Tg(gfap:nsfb-mCherry) and Tg(pu1:Gal4;UAS:rfp);Tg(gfap:nsfb-mCherry) animals stained with 4C4 showing the reduction of pu1+ microglia and no change in the mrc1a+;4C4+ microglia in the GW2580 treated animals compared to DMSO control animals. Blue arrowheads: represent mrc1a+;4C4+ microglia. Purple arrowheads represent pu1+ microglia. (K) Confocal z-projections of single pu1 cells in the embryonic yolksac of Tg(mrc1a:egfp);Tg(pu1:eos) animals at 24 hpf pre and post-photoconversion. Purple arrowheads indicate successfully photoconverted pEos+ cells. Imaging window equals 0.0027 mm3 (A-G, J-K), 0.0081 mm3 or 3000 µm (H-I). Scale bar equals 10 µm (J,K).
Extended Data Fig. 7 Injury paradigms alter mrc1a+ microglia and expression of mammalian Mrc1 during injury.
(A) Quantification of spinal cord mCherry intensity in MTZ-treated animals across time from 1 to 4 dpi compared to DMSO treated zebrafish animals (t-test, two-tailed; multiple comparisons corrected: 1 dpi DMSO vs. MTZ p < 0.0001, 2 dpi DMSO vs. MTZ p < 0.0001, 3 dpi DMSO vs. MTZ p < 0.0001, 4 dpi DMSO vs. MTZ p < 0.0001; all two-tailed)(n = 70 animals). (B) Stacked violin plot of microglia from Hammond et al. (2019) comparing canonical microglia marker gene expression between Mrc1+ and Mrc1- microglia from P100 LPC-injected animals. Microglia were subsetted as Mrc1+ if log normalized expression of Mrc1 was > 0. (C) Differential expression testing results table comparing expression of canonical microglia markers in Mrc1+ versus Mrc1- microglia from Hammond et al. (2019). In (C) “Log Fold Change” refers to natural log fold-change, with positive values indicating higher expression in Mrc1+ microglia versus Mrc1- microglia. “Mrc1 + microglia with expression” and “Mrc1- microglia with expression” report the percentage of Mrc1+ and Mrc1- cells, respectively, with at least one read of the gene detected. Statistical significance was determined by Wilcoxon rank-sum test and an average log fold-change threshold of ±0.5. Imaging window equals 0.0027 mm3 (A).
Extended Data Fig. 8 mrc1a+ microglia are dependent on lymphangiogenesis during injury.
(A) Quantification of the normalized fluorescence value of DMSO control animals compared to lymphatic inhibitor treated animals (one-way ANOVA/Dunnett;s multiple comparisons: DMSO vs. A77-1726 p = 0.997, DMSO vs. cinnarizine p = 0.4055, DMSO vs. flunarizine p = 0.0003, DMSO vs. leflunomide p = 0.0533)(n = 32 animals). (B) Representative confocal z-projections of 5 dpf Tg(mrc1a:egfp) animals showing the disruption of vessels in animals treated with A77-1726 + MTZ, cinnarizine + MTZ, or flunarizine + MT compared to control DMSO + MTZ control animals. (C) Quantification of the average length of brain lymphatic vessels in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett;s multiple comparisons: DMSO vs. A77-1726 p = 0.6526,Mean diff=1.962, DF = 44, q = 4.081, SE of diff=0.4807; DMSO vs. cinnarizine p = 0.6011, Mean diff=3.845, DF = 44, q = 3.845, SE of diff=0.5056; DMSO vs. flunarizine p = 0.0482, Mean diff=1.887, DF = 44, q = 3.265, SE of diff=0.5779; DMSO vs. leflunomide p = 0.0702, Mean diff=2.087, DF = 44, q = 3.887, SE of diff=0.5369)(n = 72 animals). (D) Quantification showing the reduced average number of secondary sprouts (Fig. S4B) in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett;s multiple comparisons: DMSO vs. A77-1726 p = 0.0007, Mean diff=3.542, DF = 45, q = 3.459, SE of diff = .024; DMSO vs. cinnarizine p = 0.0015, Mean diff=1.944, DF = 45, q = 3.845, SE of diff=0.5056; DMSO vs. flunarizine p = 0.0083, Mean diff=2.842, DF = 45, q = 2.305, SE of diff=1.233; DMSO vs. leflunomide p = 0.0014, Mean diff=3.375, DF = 45, q = 2.948, SE of diff=1.145)(n = 72 animals). (E) Quantification of the average number of lymphatic vessels that surround the brain in lymphatic inhibitor treated animals compared to DMSO control animals (one-way ANOVA/Dunnett;s multiple comparisons: DMSO vs. A77-1726 p = 0.0047, DMSO vs. cinnarizine p = 0.0271, DMSO vs. flunarizine p = 0.0953, DMSO vs. leflunomide p = 0.0196)(n = 72 animals). (F) Quantification of the number of 4C4+ only microglia in DMSO + MTZ control animals compared to leflunomide + MTZ and flunarizine + MTZ treated animals (One-way ANOVA Dunnett’s multiple comparisons: DMSO vs. leflunomide p = 0.0038, DMSO vs. flunarizine p = 0.0222)(n = 29 animals). (G) Quantification of the number of pu1+ only microglia in DMSO + MTZ control animals compared to leflunomide + MTZ and flunarizine + MTZ treated animals (One-way ANOVA Dunnett’s multiple comparisons: DMSO vs. leflunomide, DMSO vs. flunarizine p = 0.7053, Mean diff=32.76, q = 3.554, SE of diff=9.22; DMSO vs. flunarizine p = 0.5165, Mean diff=27.72, q = 2.772, SE of diff=10)(n = 29 animals). (H) Quantification of the number of pu1+;4C4+ microglia in DMSO + MTZ control animals compared to leflunomide + MTZ and flunarizine + MTZ treated animals (One-way ANOVA Dunnett’s multiple comparisons: DMSO vs. leflunomide p = 0.1400, Mean diff=1.75, q = 0.723, SE of diff=2.421; DMSO vs. flunarizine p = 0.2981, Mean diff = −2.667, q = 1.012, SE of diff=2.635) (n = 29 animals) (I) Representative confocal z-projections of 6 dpf Tg(mrc1a:egfp);Tg(gfap:nsfb-mCherry) animals stained with 4C4 showing the reduction of mrc1a+;4C4+ microglia in A77-1726 + MTZ, cinnarizine + MTZ, flunarizine + MTZ, and leflunomide + MTZ, treated animals compared to DMSO + MTZ control animals. Blue arrowheads represent mrc1a+4C4+ microglia. Imaging window equals 0.0027 mm3 (A-E,I), 3000 µm (F-H). Scale bar equals 10 µm (B,I).
Supplementary information
Supplementary Table 1
scRNA-seq differential expression results. Contains differential expression testing results for the analyses of the Hammond et al. and Kracht et al. scRNA-seq datasets. Includes results for both initial clustering to remove contaminants and subclustering, as well as results comparing Mrc1+ to Mrc1− microglia.
Supplementary Video 1
Various mrc1a+ cells interacting with and migrating around lymphatic vessels. Segment from a 24-h time-lapse movie between 48 hpf and 72 hpf showing several mrc1a+ cells migrating and interacting with lymphatic vessels in the head of zebrafish. Differently colored arrowheads indicate individual mrc1a+ cells as they interact throughout the time-lapse movie. Frame rate equals 1 frame per second (fps).
Supplementary Video 2
Segment from a 24-h time-lapse movie from 34 hpf to 58 hpf of a migrating mrc1a+ cell. Contains frames from a 24-h time-lapse movie of Tg(mrc1a:gfp) animals showing an mrc1a+ cell exiting and encircling a lymphatic vessel surrounding the zebrafish brain. Frame rate equals 1 frame per second (fps). Arrowhead (top) denotes the migrating mrc1a+ cell and the line (bottom) denotes the path of migration.
Supplementary Video 3
Segment from a 24-h time-lapse movie from 34 hpf to 58 hpf from Supplementary Video 2. Movie is an IMARIS surface reconstruction of an mrc1a+ cell exiting and encircling a lymphatic vessel surrounding the zebrafish brain. Frame rate equals 1 frame per second (fps).
Source data
Source Data Fig. 1
All raw n values, data values and statistical data exported from GraphPad Prism for Fig. 1.
Source Data Fig. 2
All raw n values, data values and statistical data exported from GraphPad Prism for Fig. 2.
Source Data Fig. 3
All raw n values, data values and statistical data exported from GraphPad Prism for Fig. 3.
Source Data Fig. 4
All raw n values, data values and statistical data exported from GraphPad Prism for Fig. 4.
Source Data Fig. 5
All raw n values, data values and statistical data exported from GraphPad Prism for Fig. 5.
Source Data Fig. 6
All raw n values, data values and statistical data exported from GraphPad Prism for Fig. 6.
Source Data Fig. 7
All raw n values, data values and statistical data exported from GraphPad Prism for Fig. 7.
Source Data Fig. 8
All raw n values, data values and statistical data exported from GraphPad Prism for Fig. 8.
Source Data Extended Data Fig. 1
All raw n values, data values and statistical data exported from GraphPad Prism for Extended Data Fig. 1.
Source Data Extended Data Fig. 2
All raw n values, data values and statistical data exported from GraphPad Prism for Extended Data Fig. 2.
Source Data Extended Data Fig. 3
All raw n values, data values and statistical data exported from GraphPad Prism for Extended Data Fig. 3.
Source Data Extended Data Fig. 4
All raw n values, data values and statistical data exported from GraphPad Prism for Extended Data Fig. 4.
Source Data Extended Data Fig. 5
All raw n values, data values and statistical data exported from GraphPad Prism for Extended Data Fig. 5.
Source Data Extended Data Fig. 6
All raw n values, data values and statistical data exported from GraphPad Prism for Extended Data Fig. 6.
Source Data Extended Data Fig. 7
All raw n values, data values and statistical data exported from GraphPad Prism for Extended Data Fig. 7.
Source Data Extended Data Fig. 8
All raw n values, data values and statistical data exported from GraphPad Prism for Extended Data Fig. 8.
Rights and permissions
About this article
Cite this article
Green, L.A., O’Dea, M.R., Hoover, C.A. et al. The embryonic zebrafish brain is seeded by a lymphatic-dependent population of mrc1+ microglia precursors. Nat Neurosci 25, 849–864 (2022). https://doi.org/10.1038/s41593-022-01091-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01091-9