Abstract
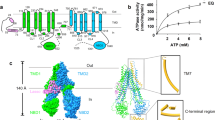
Multidrug resistance protein 4 (MRP4) is a broadly expressed ATP-binding cassette transporter that is unique among the MRP subfamily for transporting prostanoids, a group of signaling molecules derived from unsaturated fatty acids. To better understand the basis of the substrate selectivity of MRP4, we used cryogenic-electron microscopy to determine six structures of nanodisc-reconstituted MRP4 at various stages throughout its transport cycle. Substrate-bound structures of MRP4 in complex with PGE1, PGE2 and the sulfonated-sterol DHEA-S reveal a common binding site that accommodates a diverse set of organic anions and suggest an allosteric mechanism for substrate-induced enhancement of MRP4 ATPase activity. Our structure of a catalytically compromised MRP4 mutant bound to ATP-Mg2+ is outward-occluded, a conformation previously unobserved in the MRP subfamily and consistent with an alternating-access transport mechanism. Our study provides insights into the endogenous function of this versatile efflux transporter and establishes a basis for MRP4-targeted drug design.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All five three-dimensional cryo-EM density maps have been deposited to the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-40829 (apowide MRP4), EMD-40828 (aponarrow MRP4), EMD-40827 (PGE1-bound MRP4), EMD-40830 (PGE2-bound MRP4), EMD-40826 (DHEA-S-bound MRP4) and EMD-40821 (ATP-bound MRP4E1202Q). The coordinates for the atomic models have been deposited in the Protein Data Bank (PDB) under accession numbers 8SXA (apowide MRP4), 8SX9 (aponarrow MRP4), 8SX8 (PGE1-bound MRP4), 8SXB (PGE2-bound MRP4), 8SX7 (DHEA-S-bound MRP4) and 8SWN (ATP-bound MRP4E1202Q). See Table 1 for more details. Source data are provided with this paper.
References
Chabowski, D. S., Cohen, K. E., Abu-Hatoum, O., Gutterman, D. D. & Freed, J. K. Crossing signals: bioactive lipids in the microvasculature. Am. J. Physiol. Heart Circ. Physiol. 318, H1185–H1197 (2020).
Harizi, H. & Gualde, N. The impact of eicosanoids on the crosstalk between innate and adaptive immunity: the key roles of dendritic cells. Tissue Antigens 65, 507–514 (2005).
Martens, M. D., Fernando, A. S. & Gordon, J. W. A new trick for an old dog? Myocardial-specific roles for prostaglandins as mediators of ischemic injury and repair. Am. J. Physiol. Heart Circ. Physiol. 320, H2169–H2184 (2021).
Mitchell, J. A. & Kirkby, N. S. Eicosanoids, prostacyclin and cyclooxygenase in the cardiovascular system. Br. J. Pharmacol. 176, 1038–1050 (2019).
Oyesola, O. O. & Tait Wojno, E. D. Prostaglandin regulation of type 2 inflammation: from basic biology to therapeutic interventions. Eur. J. Immunol. 51, 2399–2416 (2021).
Vannuccini, S., Bocchi, C., Severi, F. M., Challis, J. R. & Petraglia, F. Endocrinology of human parturition. Ann. Endocrinol. 77, 105–113 (2016).
Zeilhofer, H. U. Prostanoids in nociception and pain. Biochem. Pharmacol. 73, 165–174 (2007).
Biringer, R. G. The enzymology of the human prostanoid pathway. Mol. Biol. Rep. 47, 4569–4586 (2020).
Johnson, A. M., Kleczko, E. K. & Nemenoff, R. A. Eicosanoids in cancer: new roles in immunoregulation. Front Pharm. 11, 595498 (2020).
Mitchell, J. A. et al. Cyclooxygenases and the cardiovascular system. Pharmacol. Ther. 217, 107624 (2021).
Wang, D., Cabalag, C. S., Clemons, N. J. & DuBois, R. N. Cyclooxygenases and prostaglandins in tumor immunology and microenvironment of gastrointestinal cancer. Gastroenterology 161, 1813–1829 (2021).
Beaman, J., Prifti, C., Schwarz, E. B. & Sobota, M. Medication to manage abortion and miscarriage. J. Gen. Intern Med 35, 2398–2405 (2020).
Klimko, P. G. & Sharif, N. A. Discovery, characterization and clinical utility of prostaglandin agonists for the treatment of glaucoma. Br. J. Pharmacol. 176, 1051–1058 (2019).
Lang, I. M. & Gaine, S. P. Recent advances in targeting the prostacyclin pathway in pulmonary arterial hypertension. Eur. Respir. Rev. 24, 630–641 (2015).
Lee, O. Y. et al. A comparative study of DA-9601 and misoprostol for prevention of NSAID-associated gastroduodenal injury in patients undergoing chronic NSAID treatment. Arch. Pharm. Res 37, 1308–1316 (2014).
Stephenson, M. L. & Wing, D. A. Misoprostol for induction of labor. Semin Perinatol. 39, 459–462 (2015).
Reid, G. et al. The human multidrug resistance protein MRP4 functions as a prostaglandin efflux transporter and is inhibited by nonsteroidal antiinflammatory drugs. Proc. Natl Acad. Sci. USA 100, 9244–9249 (2003).
Wen, J. et al. The pharmacological and physiological role of multidrug-resistant protein 4. J. Pharmacol. Exp. Ther. 354, 358–375 (2015).
Rius, M., Thon, W. F., Keppler, D. & Nies, A. T. Prostanoid transport by multidrug resistance protein 4 (MRP4/ABCC4) localized in tissues of the human urogenital tract. J. Urol. 174, 2409–2414 (2005).
Tanaka, N., Kawai, J., Hirasawa, N., Mano, N. & Yamaguchi, H. ATP-Binding cassette transporter C4 is a prostaglandin D2 exporter in HMC-1 cells. Prostaglandins Leukot. Ess. Fat. Acids 159, 102139 (2020).
Kalinski, P. Regulation of immune responses by prostaglandin E2. J. Immunol. 188, 21–28 (2012).
Kochel, T. J., Reader, J. C., Ma, X., Kundu, N. & Fulton, A. M. Multiple drug resistance-associated protein (MRP4) exports prostaglandin E2 (PGE2) and contributes to metastasis in basal/triple negative breast cancer. Oncotarget 8, 6540–6554 (2017).
Zelcer, N. et al. Steroid and bile acid conjugates are substrates of human multidrug-resistance protein (MRP) 4 (ATP-binding cassette C4). Biochem. J. 371, 361–367 (2003).
Chen, Z. S., Lee, K. & Kruh, G. D. Transport of cyclic nucleotides and estradiol 17-b-d-glucuronide by multidrug resistance protein 4. Resistance to 6-mercaptopurine and 6-thioguanine. J. Biol. Chem. 276, 33747–33754 (2001).
Hara, Y. et al. Inhibition of MRP4 prevents and reverses pulmonary hypertension in mice. J. Clin. Invest. 121, 2888–2897 (2011).
Decouture, B. et al. Impaired platelet activation and cAMP homeostasis in MRP4-deficient mice. Blood 126, 1823–1830 (2015).
Li, C. et al. Spatiotemporal coupling of cAMP transporter to CFTR chloride channel function in the gut epithelia. Cell 131, 940–951 (2007).
Moon, C. et al. Compartmentalized accumulation of cAMP near complexes of multidrug resistance protein 4 (MRP4) and cystic fibrosis transmembrane conductance regulator (CFTR) contributes to drug-induced diarrhea. J. Biol. Chem. 290, 11246–11257 (2015).
Chen, Z. S. et al. Analysis of methotrexate and folate transport by multidrug resistance protein 4 (ABCC4): MRP4 is a component of the methotrexate efflux system. Cancer Res. 62, 3144–3150 (2002).
Ci, L. et al. Involvement of MRP4 (ABCC4) in the luminal efflux of ceftizoxime and cefazolin in the kidney. Mol. Pharmacol. 71, 1591–1597 (2007).
Imaoka, T. et al. Functional involvement of multidrug resistance-associated protein 4 (MRP4/ABCC4) in the renal elimination of the antiviral drugs adefovir and tenofovir. Mol. Pharmacol. 71, 619–627 (2007).
Kohler, J. J. et al. Tenofovir renal proximal tubular toxicity is regulated by OAT1 and MRP4 transporters. Lab. Invest. 91, 852–858 (2011).
Ray, A. S. et al. Mechanism of active renal tubular efflux of tenofovir. Antimicrob. Agents Chemother. 50, 3297–3304 (2006).
Rius, M., Hummel-Eisenbeiss, J., Hofmann, A. F. & Keppler, D. Substrate specificity of human ABCC4 (MRP4)-mediated cotransport of bile acids and reduced glutathione. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G640–G649 (2006).
Rius, M., Hummel-Eisenbeiss, J. & Keppler, D. ATP-dependent transport of leukotrienes B4 and C4 by the multidrug resistance protein ABCC4 (MRP4). J. Pharmacol. Exp. Ther. 324, 86–94 (2008).
Rius, M., Nies, A. T., Hummel-Eisenbeiss, J., Jedlitschky, G. & Keppler, D. Cotransport of reduced glutathione with bile salts by MRP4 (ABCC4) localized to the basolateral hepatocyte membrane. Hepatology 38, 374–384 (2003).
Vogt, K. et al. Release of platelet-derived sphingosine-1-phosphate involves multidrug resistance protein 4 (MRP4/ABCC4) and Is inhibited by statins. Thromb. Haemost. 118, 132–142 (2018).
Hagmann, W. et al. Purification of the human apical conjugate export pump MRP2 reconstitution and functional characterization as substrate-stimulated ATPase. Eur. J. Biochem. 265, 281–289 (1999).
Mao, Q., Leslie, E. M., Deeley, R. G. & Cole, S. P. ATPase activity of purified and reconstituted multidrug resistance protein MRP1 from drug-selected H69AR cells. Biochim. Biophys. Acta 1461, 69–82 (1999).
Shapiro, A. B. & Ling, V. ATPase activity of purified and reconstituted P-glycoprotein from Chinese hamster ovary cells. J. Biol. Chem. 269, 3745–3754 (1994).
Sauna, Z. E., Nandigama, K. & Ambudkar, S. V. Multidrug resistance protein 4 (ABCC4)-mediated ATP hydrolysis: effect of transport substrates and characterization of the post-hydrolysis transition state. J. Biol. Chem. 279, 48855–48864 (2004).
Berthier, J., Arnion, H., Saint-Marcoux, F. & Picard, N. Multidrug resistance-associated protein 4 in pharmacology: overview of its contribution to pharmacokinetics, pharmacodynamics and pharmacogenetics. Life Sci. 231, 116540 (2019).
Stockner, T., Gradisch, R. & Schmitt, L. The role of the degenerate nucleotide binding site in type I ABC exporters. FEBS Lett. 594, 3815–3838 (2020).
Copsel, S. et al. Multidrug resistance protein 4 (MRP4/ABCC4) regulates cAMP cellular levels and controls human leukemia cell proliferation and differentiation. J. Biol. Chem. 286, 6979–6988 (2011).
Hayashi, H. et al. Sorting nexin 27 interacts with multidrug resistance-associated protein 4 (MRP4) and mediates internalization of MRP4. J. Biol. Chem. 287, 15054–15065 (2012).
Payen, L. F., Gao, M., Westlake, C. J., Cole, S. P. & Deeley, R. G. Role of carboxylate residues adjacent to the conserved core Walker B motifs in the catalytic cycle of multidrug resistance protein 1 (ABCC1). J. Biol. Chem. 278, 38537–38547 (2003).
Johnson, Z. L. & Chen, J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085 e9 (2017).
Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017).
Hofmann, S. et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019).
Sedzicki, J. et al. Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella. Nat. Struct. Mol. Biol. 29, 1170–1177 (2022).
Choudhury, H. G. et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl Acad. Sci. USA 111, 9145–9150 (2014).
Olsen, J. A., Alam, A., Kowal, J., Stieger, B. & Locher, K. P. Structure of the human lipid exporter ABCB4 in a lipid environment. Nat. Struct. Mol. Biol. 27, 62–70 (2020).
Lee, K. P. K., Chen, J. & MacKinnon, R. Molecular structure of human KATP in complex with ATP and ADP. eLife 6, e32481 (2017).
El-Sheikh, A. A., van den Heuvel, J. J., Krieger, E., Russel, F. G. & Koenderink, J. B. Functional role of arginine 375 in transmembrane helix 6 of multidrug resistance protein 4 (MRP4/ABCC4). Mol. Pharmacol. 74, 964–971 (2008).
Wittgen, H. G. et al. Phenylalanine 368 of multidrug resistance-associated protein 4 (MRP4/ABCC4) plays a crucial role in substrate-specific transport activity. Biochem. Pharmacol. 84, 366–373 (2012).
Schuetz, J. D. et al. MRP4: a previously unidentified factor in resistance to nucleoside-based antiviral drugs. Nat. Med. 5, 1048–1051 (1999).
van Aubel, R. A., Smeets, P. H., Peters, J. G., Bindels, R. J. & Russel, F. G. The MRP4/ABCC4 gene encodes a novel apical organic anion transporter in human kidney proximal tubules: putative efflux pump for urinary cAMP and cGMP. J. Am. Soc. Nephrol. 13, 595–603 (2002).
Kawahara, K., Hohjoh, H., Inazumi, T., Tsuchiya, S. & Sugimoto, Y. Prostaglandin E2-induced inflammation: relevance of prostaglandin E receptors. Biochim. Biophys. Acta 1851, 414–421 (2015).
Barnett, R. E. Effect of monovalent cations on the ouabain iniibition of the sodium and potassium ion activated adenosine triphosphatase. Biochemistry 9, 4644–4648 (1970).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pravda, L. et al. MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 46, W368–W373 (2018).
Voss, N. R. & Gerstein, M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 38, W555–W562 (2010).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Pintilie, G. et al. Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods 17, 328–334 (2020).
Acknowledgements
We thank P. Nguyen for assistance with virus production and D. Asarnow for discussions regarding purification and reconstitution of MRP4. This work was supported by the National Institutes of Health (NIH) National Institute of General Medical Sciences (grants NIGMS GM24485 to R.M.S. and R35GM140847 to Y.C.). R.B. was supported in part by American Heart Association postdoctoral fellowship award 9POST34370101. Mass spectrometry was performed in the UCSF Mass Spectrometry facility supported by NIH grant P41 GM103481. We thank D. P. Bulkley, G. Gilbert and M. Harrington for support with cryo-EM data collection. All data were collected at the UCSF cryo-EM facility, which is supported by NIH grants S10OD020054, S10OD021741 and S10OD026881.
Author information
Authors and Affiliations
Contributions
R.M.S., D.L.K., C.S.C., Y.C. and A.S. conceived this work. S.P., R.B. and G.M.K. expressed and functionally characterized proteins. S.P., E.G. and M.G. collected and processed cryo-EM data. S.P. and E.G. performed model building and refinement. S.P., R.B., E.G., I.E.C., R.M.S. and D.L.K. wrote the manuscript with contributions from all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Kenneth Linton and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sequence alignment of MRP4 and MRP1 homologs.
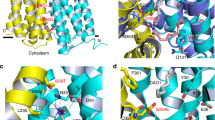
Sequences of bovine and human MRP4 are aligned to reflect sequence conservation across evolution and similarity to the closely related MRP1. All references to MRP4 used throughout the text refer to the bovine sequence. The residue numbers for bovine MRP4 are indicated above the sequences. Red asterisks denote a residue involved in binding one of either DHEA-S, PGE1, or PGE2. Text boxes refer to conserved sequence motifs from NBD1 (pink) and NBD2 (cyan), NBD2’s inserted sequence (orange) and the C-terminal PDZ-binding motif (PDZ-BM, sand). Bovine MRP4 catalytic residue E1202 is highlighted in red.
Extended Data Fig. 2 Biochemical characterization of MRP4.
Fold change in ATPase activity relative to basal levels with the addition of increasing concentrations of a, PGE1, PGE2, b, DHEAS, c, cAMP, cGMP, d, LTC4, e, tenofovir, f, E217G, g, folic acid, h, S1P. Data in each panel were obtained from separate preparations of MRP4 in MSP lipid nanodisc. The basal ATPase specific activity across the samples used in panels a-f was 13.6 ± 4.5 nmol min-1 mg-1 from 10 biological replicates. PGE1, prostaglandin E1; PGE2, prostaglandin E2; cAMP, cyclic adenosine monophosphate; cGMP, cyclic guanosine monophosphate; LTC4, leukotriene C4; DHEA-S, dehydroepiandrosterone sulfate; E217G, β-estradiol-17β-D-glucuronide; S1P, sphingosine-1-phosphate.
Extended Data Fig. 3 cAMP does not elicit a conformational change in MRP4 in cryo-EM.
a Final density of MRP4 in the presence of 1 mM cAMP from cryoSPARC. Sharpened output volume in gray, lowpass filtered volume showing the nanodisc and NBDs as black silhouette. b Rigid body fitting of apo MRP4 into density obtained in the presence of 1 mM cAMP. Density in gray, MRP4 domains colored as before. c View of substrate binding residues of the rigid-body fit apo MRP4 model in the density obtained for MRP4 in the presence of 1 mM cAMP. Map contoured at low threshold, revealing non-protein density similar to our apo MRP4 refinement. Density in purple mesh, MRP4 domains colored as before.
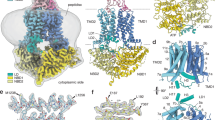
Extended Data Fig. 4 The domains of MRP4 move as rigid bodies throughout the substrate transport cycle.
Superposition of a, bundle 1 (TMs 1, 2, 3, 6, 10, 11) b, bundle 2 (TMs 4, 5, 7, 8, 9, 12) c, NBD1 and d, NBD2 across all five structures. Apowide MRP4 in salmon, aponarrow MRP4 in green, DHEA-S-bound MRP4 in cyan, PGE1-bound MRP4 in grey, PGE2-bound MRP4 in pink, ATP-Mg2+-bound MRP4 in yellow. e, Superposition of the three substrate-bound structures reveals them to share an inward-open, narrow conformation. RMSD between all Cα of DHEA-S-bound MRP4 and PGE1-bound MRP4 is 1.2 Å; between DHEA-S-bound and PGE2-bound is 0.87 Å; between PGE1-bound and PGE2-bound is 0.90 Å. Structures are colored as in a-d.
Extended Data Fig. 5 The outward-facing occluded state of MRP4 closely resembles the structure of SUR1 bound to ADP/ATP.
a Front view, b side view, and c top view of MRP4E1202Q bound to ATP-Mg2+ and SUR1 bound to ADP-ATP (shown as single chain from PDB ID:6C3O). TMD0 domain in SUR1 is hidden for clarity in a and c. Cα RMSD between our ATP-bound structure and SUR1 with TMD0 domain deleted is 4.36 Å. MRP4E1202Q bound to ATP-Mg2+ in pink, SUR1 bound to ADP-ATP in cyan, TMD0 domain in light gray.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6.
Source data
Source Data Fig. 1
SEC A280 source data for Fig. 1b; Spectromax A340 source data for Fig. 1c,d,e.
Source Data Fig. 2
Uncropped gel image for Fig. 1b. Coomassie brilliant blue stained gel. Alternating lanes of ladders and serial dilutions of the peak MRP4–nanodisc fraction from SEC.
Source Data Extended Data Fig. 2
Spectromax A340 source data for Extended Fig. 2.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pourmal, S., Green, E., Bajaj, R. et al. Structural basis of prostaglandin efflux by MRP4. Nat Struct Mol Biol 31, 621–632 (2024). https://doi.org/10.1038/s41594-023-01176-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-01176-4